Research Scholar ,Regenix Super Speciality laboratories Pvt.Ltd. (Affliated to University of Madras ).No.42 ,
Loganathan Nagar , 100 feet road, Choolaimedu (Vadapalani),Chennai -600094,Tamil Nadu ,India.
1,2 and 4 -Regenix Super Speciality laboratories pvt.ltd.
3- Apollo Lab Services
Corresponding author email: poornimashyambt@gmail.com
Article Publishing History
Received: 15/04/2023
Accepted After Revision: 28/06/2023
Bacteria are one of the most diverse organisms that exist. Every day labs are flooded with cultures to detect the type of microorganisms and their antibiotic resistance patterns to treat patients. However, this reporting takes 3 days. Molecular biology is a field that is associated with quicker reporting. This work is an attempt to make molecular testing easier and quicker to help with one step DNA extraction method. 3 different buffers were tested for DNA extraction from cultures. The result of the best buffer is represented in agarose gel electrophoresis. Efficient extraction of DNA from bacteria was seen. The first step to standardization of buffers for the one-step extraction of DNA from bacteria. The buffers need to be tested and standardized with a wider variety of samples of both gram-positive and gram-negative organisms and an efficient method of extraction from the primary samples instead of the culture plates has to be standardized. This is a pilot study to screen the buffers for their effectiveness.
bacterial DNA, Gram-positive, Gram Negative, buffers, Agarose gel electrophoresis.
Shyam P, Dr. Subramaniam S, Dr. Subramaniam S, Jospiene M E.. Standardization and Evaluation of Buffers: A One Step DNA Extraction Protocol from Microbial Cultures. Biosc.Biotech.Res.Comm. 2023;16(2).
Shyam P, Dr. Subramaniam S, Dr. Subramaniam S, Jospiene M E. Standardization and Evaluation of Buffers: A One Step DNA
Extraction Protocol from Microbial Cultures. Biosc.Biotech.Res.Comm. 2023;16(2). Available from: <a href=”https://bit.ly/2U8EBeg“>https://bit.ly/2U8EBeg</a>
INTRODUCTION
Infection by microorganisms is an overwhelming scenario that causes serious implications in the patients and sometimes even causes death. Bacteria are a major cause of these infections. Bacteria are defined as a group of small, single-celled organisms. The extent of damage is measured by the route, number, mode of transmission, and stability of the host. While the nose, throat, and cuts in the skin are foregrounds of microbial infection, the lungs, bowel, urinary tract, vagina, etc are also sites of serious infections, ( Cars & Nordberg 2005, Weber & Stilianakis 2008 Reynolds & Kollef 2021).
The major mode of infection is entry through the skin and replication in the tissue. The incidence of infectious diseases was estimated at 71.8 per 10,000 population in 2022. These infectious diseases kill about nine million individuals per year among which children under the age of 5 are a major part. Bacterial diseases causing infection have reached various advanced levels of testing. Techniques of Lateral flow assay, ELISA, and molecular testing techniques have been employed (Lamballerie et al., 1992, Boehringer and O’Farrell, 2021).
However, for infections in wounds, the techniques of microbial identification and consequently identification of the antibiotics to treat the infection is a 3-day process. The samples for anti-microbial testing are in the order of sputum, urine, high vaginal swab, wound swab, and finally blood. Anti-microbial resistance due to prolonged use of antibiotics is one of the biggest challenges of the microbial industry today. Some of the major bacteria that cause infections are Klebsiella spp. , Moraxella spp.,Escherichia spp., Pseudomonas spp. , Staphylococcus spp. etc., (Bharathi et al., 2010, Reynolds & Kollef 2021).
The constant evolution of bacteria poses a major challenge in identification. Testing like miRNA and SNP may be an apt solution (Ali et al., 2017). The bacterial cell wall is made up of rigid structures of uniform thickness around the cell. This rigidness ensures the various types of shapes like spiral, rod, and coccus and patterns of colonization. Based on cell wall components bacteria are divided into Gram-positive and Gram-negative bacteria (Schleifer et al., 1972). The presence of a peptidoglycan layer in the outer layer of the Gram-negative bacteria makes the need for the buffer to lyse the cell wall different from that of the gram-positive bacteria.
This study deals with the standardization of a buffer that extracts bacterial DNA from overnight culture plates in one step. The purity of the DNA obtained was screened with A 260/ 280 and confirmed with Agarose gel electrophoresis. The results displayed below are of the best methods that yielded a good quantity of DNA.
Materials and Methods: The bacteria for testing was taken from ATCC strains. The confirmation was done by staining and biochemical studies. One gram- Positive and one gram-negative bacteria were taken for study.Gram Positive – Staphylococcus aureus– ATCC @ 25923 Gram Negative – Escherichia coli ATCC 25922. The same was inoculated for overnight growth in nutrient broth. One ml of broth was taken centrifuged and used for the rest of the experiment.
Buffers: The below table contains the constituents of the best 3 buffers which were taken for further studies.
Table 1. buffers and their ingredients
| Buffer 1:
Tris HCL KCL (Pottasium chloride ) Mgcl2(Magnesium chloride) SDS 2% (Sodium Dodecyl Sulfate) Triton X Ethanol |
Buffer 2:
MgCl2 EDTA SDS Triton X100 PVP (Polyvinylpyrrolidone) CH3COOK TE buffer (Tris –EDTA) Ethanol |
Buffer3:
Tris HCL TE buffer MgCl2 EDTA SDS PVP CH3COOK Ethanol NaCl(Sodium Chloride) |
Absorbance at 260 nm, 280nm, and 230nm.:NanoDrop® ND-1000 was used to take the readings (5). Quantification of DNA : dsDNA concentration = 50 μg/mL × OD260 × dilution factor
Gel Electrophoresis: A 1% gel was used for the analysis of the result. Only pure DNA was used for this analysis. The gel pictures of the best results are represented in the results section. The running buffer of TAE and TBE was used. To visualize DNA EtBr (Ethidium Bromide) was added and visualized through a UV transilluminator and documented through a gel documentation system (Lee et al., 2012).
RESULTS AND DISCUSSION
All experiments have been carried out in triplicate. The below results are the representatives of the same.The purity of the DNA obtained is seen from the A260/280 ratio. A value between 1.7 -2.0 is considered pure for ds DNA. A lower value indicated the presence of phenol, proteins, and other contaminants as they are easily absorbed at 280nm. A higher value of the ratio shows the presence of RNase. In this experiment as proteinase K is not used to avoid the barrier of storage the need to rule out contamination becomes important. DNA Purity is calculated using the below formula and represented in the table (Ning et al., 2009).
DNA Purity (A260/A280) = (A260 reading – A320 reading) ÷ (A280 reading – A320 reading) (Tiwari et al., 2017).. A260/230 is needed to rule out contaminants such as guanidine thiocyanate, guanidine HCL, triazole, phenol, etc. A ratio between 2.0 to 2.2 is considered pure for this ratio.The below table contains the results of the nanodrop for the evaluation of the buffers: The results of the effective buffers are only represented in the table. buffers and methods which showed values of ratios below 1.5 or above 2.2 for ratios A260/A280 are not represented.
Table 2. Nanodrop reading of the DNA extact and quantification
| S.no. | Buffer name | Method | A260/280 | A2260/230 | DNA Quantification ng/μl |
| 1. | Buffer 1 | A | 1.68 | 2.2 | 4.1 |
| B | 1.75 | 2.19 | 8.3 | ||
| 2. | Buffer 2 | A | 1.85 | 2.18 | 27.2 |
| B | 1.71 | 2.1 | 10.7 | ||
| 3. | Buffer 3 | A | 1.72 | 2.19 | 11.2 |
| B | 1.91 | 2.02 | 33.4 |
A-Staphylococcus aureus B- Escherichia coli
This result shows that buffer 2 is more efficient for S. aureus – gram-positive, while buffer 3 is more efficient for E.coli – gram-negative. This shows PVP and CH3COOK increase the efficiency of extraction. CH3COOK helps with the breaking down of proteins ( Angela 2017). PVP increases the DNA yield (Khan et al., 2007). Buffer 2 with the second detergent, Triton X is more efficient in the case of Gram –Positive bacteria. While Tris HCL and Nacl proved greater efficiency in the breakdown of the peptidoglycan layer presenting Gram Negative bacteria. Tris HCL helps in the maintenance of the buffer layer whereas NaCl proves efficient with the lysis of the peptidoglycans by creating pores on the surface of the cell wall.
Gel electrophoresis.
Figure 1: Lane 1 – ladder Lane 2 and 3, Buffer 3 – Escherichia coli. Lane 4 and 5 , Buffer2 – Staphylococcus aureus
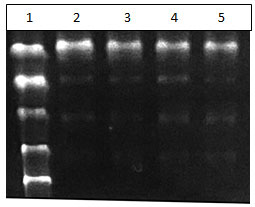
Separation of DNA is seen. Addition of Ethanol to collect the DNA yields pure DNA that can be used for further study.
Figure 2 : DNA after ethanol wash. Lane 1: DNA Ladder Lane 2 : gram positive bacteria from kit method. Lane 3 : gram negative bacteria from kit method . Lane 4 and 5- Gram negative bacteria from buffer 3. Lane 6 and 7 Gram positive bacteria from buffer 2.
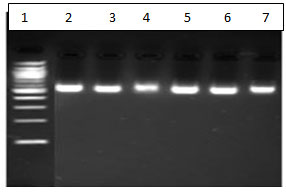
Efficient extraction comparable to a CE-approved extraction kit was seen.
CONCLUSION
Efficient extraction of DNA from bacteria was seen. The first step to standardization of buffers for the one-step extraction of DNA from bacteria. The buffers need to be tested and standardized with a wider variety of samples of both gram-positive and gram-negative organisms and an efficient method of extraction from the primary samples instead of the culture plates has to be standardized. This is a pilot study to screen the buffers for their effectiveness.
Conflict of Interest: Authors have no conflict of interest.
REFERENCES
Ali, N., Rampazzo, R. C. P., Costa, A. D. T., & Krieger, M. A. (2017). Current Nucleic Acid Extraction Methods and Their Implications to Point-of-Care Diagnostics. BioMed research international, 2017, 9306564.
Bharathi, M. J., Ramakrishnan, R., Shivakumar, C., Meenakshi, R., & Lionalraj, D. (2010). Etiology and antibacterial susceptibility pattern of community-acquired bacterial ocular infections in a tertiary eye care hospital in south India. Indian journal of ophthalmology, 58(6), 497-507.
Boehringer HR and Brendan J O’Farrell (2021) Lateral Flow Assays in Infectious Disease Diagnosis Clinical Chem. 2021 Dec 30;68(1):52-58. doi: 10.1093/clinchem/hvab194.
Cars, O., & Nordberg, P. (2005). Antibiotic resistance–The faceless threat. International Journal of Risk & Safety in Medicine, 17(3-4), 103-110.
De Lamballerie, X., Zandotti, C., Vignoli, C., Bollet, C., & De Micco, P. (1992). A one-step microbial DNA extraction method using “Chelex 100” suitable for gene amplification. Research in microbiology, 143(8), 785-790.
Desjardins, P., & Conklin, D. (2010). NanoDrop microvolume quantitation of nucleic acids. Journal of visualized experiments : JoVE, (45), 2565.
Angel DS (2017). Measuring Elasticity of Force sensing proteins using Single Molecule Force Spectroscopy (Doctoral dissertation, IISER-M).
Khan, S., Qureshi, M. I., Alam, T., & Abdin, M. Z. (2007). Protocol for isolation of genomic DNA from dry and fresh roots of medicinal plants suitable for RAPD and restriction digestion. African Journal of Biotechnology, 6(3), 175.
Lee, P. Y., Costumbrado, J., Hsu, C. Y., & Kim, Y. H. (2012). Agarose gel electrophoresis for the separation of DNA fragments. Journal of visualized experiments : JoVE, (62), 3923.
Ning, J., Liebich, J., Kästner, M., Zhou, J., Schäffer, A., & Burauel, P. (2009). Different influences of DNA purity indices and quantity on PCR-based DGGE and functional gene microarray in soil microbial community study. Applied Microbiology and Biotechnology, 82, 983-993.
Reynolds D & M Kollef (2021) The Epidemiology and Pathogenesis and Treatment of Pseudomonas aeruginosa Infections: An Update Drugs volume 81, pages2117–2131
Schleifer, K. H., & Kandler, O. (1972). Peptidoglycan types of bacterial cell walls and their taxonomic implications. Bacteriological reviews, 36(4), 407-477.
Tiwari, S., Tomar, R. S., Tripathi, M. K., & Ahuja, A. (2017). Modified protocol for plant genomic DNA isolation. Indian Research Journal of Genetics and Biotechnology, 9(4), 478-485.
Weber, T. P., & Stilianakis, N. I. (2008). Inactivation of influenza A viruses in the environment and modes of transmission: a critical review. Journal of infection, 57(5), 361-373.


