1Department of Pediatrics, Kashan University of Medical Sciences, Kashan
2Anatomical Sciences Research Center, Kashan University of Medical Sciences, Kashan
Article Publishing History
Received: 11/11/2016
Accepted After Revision: 26/12/2016
We have investigated the correlation of multi-drug resistance- c.3435T>C common gene transition with breast cancer risk in Asian populations by an updated meta-analysis which followed by an in silico approach. In a meta-analysis approach, we collected all related studies. For this purpose, we used common electronic databases such as PubMed, Google Scholar and Science Direct. We employed bioinformatics to evaluate molecular effects of c.3435T>C transition. Our data revealed that there is no significant association between the polymorphism and breast cancer within Asian population. But, stratified meta-analysis revealed that there are a significant associations within T vs. C (OR=1.40, 95%CI=1.14-1.73, P=0.002), TT vs. CC (OR=1.85, 95%CI=1.15-2.98, P=0.011), and TT vs. CC+CT (OR=1.59, 95%CI=1.06-2.37, P=0.024) genetic models. Also, bioinformatics data revealed that c.3435T>C polymorphism could affect splicing pattern. Based on results, the c.3435T>C transition with effects on RNA splicing pattern is a risk factor for breast cancer in Iranian populations.
Breast Cancer, Genetic Polymorphism, Risk Factor, Meta-Analysis
Kheirkhah D, Karimian M. A Common Transition in Multi-Drug Resistance Gene and Risk of Breast Cancer: A Genetic Association Study with an in Silico-Analysis. Biosc.Biotech.Res.Comm. 2016;9(4).
Kheirkhah D, Karimian M. A Common Transition in Multi-Drug Resistance Gene and Risk of Breast Cancer: A Genetic Association Study with an in Silico-Analysis. Biosc.Biotech.Res.Comm. 2016;9(4). Available from: https://bit.ly/2q26J3L
Introduction
Breast cancer is the most prevalent cancer in women all over the world, and its occurrence is rising. It is a multifactorial disease that is caused by complicated interactions among genetic and environmental factors, (Jemal et al., 2006 and Nickels et al., 2013). Lately, a growing amount of research have been focused to assess the association among genetic factors and breast cancer susceptibility. Moreover, individual genetic variations, especially genetic polymorphisms in enzymes that metabolize drugs have an important role in metabolism and the fate of drugs. Also, some studies have showed that genetic polymorphisms of the multi-drug resistance1 (MDR1; ABCB1) gene, may be correlated with cancer risk (Turgut et al., 2007). The MDR1 gene that encodes a membrane-bound phosphoglycoprotein (P-gp), is positioned on chromosome 7 with 29 exons. It functions as an efflux pump which protects the cells against numerous elements including amino acids, proteins, organic cations, and others, (Kreile et al., 2013, Ozdemir et al., 2013, Lee, 2016 and Isvoran et al., 2016 ).
More than 50 variants exist in the MDR1 gene. Among these, three following single nucleotide polymorphisms (SNPs) are more common than others: (i) C1236T (rs1128503), (ii) G2677T (rs2032582), and (iii) C3435T (rs1045642; c.3435T>C). Some previous reports reported that c.3435T>C single nucleotide polymorphism in MDR1 gene may contribute in breast cancer risk (Johnatty et al., 2013 and Tazzite et al., 2016).
In this study, we have investigated the association of aforementioned transition with breast cancer risk within Asian populations by a meta-analysis and in silico approach.
Material And Methods
The literature search was done by both two authors of this paper (DK and MK). Standard electronic databases such as PubMed, Google Scholar, and Science Direct were explored for eligible papers up to Dec, 2016. Keywords and phrases which used for our search strategy were as follow: (“multi-drug resistance” or “MDR” or “ABCB1” or “C3435T” or “rs1045642”), and (“genetic polymorphism” or “SNP” or “variant” or “single nucleotide polymorphism”) and (“breast cancer”). Citations in possible eligible papers were also studied as a subsidiary source to recognize more eligible papers.
Included studies in the meta-analysis were chosen based on the following criteria: (1) the paper evaluated the association between the MDR1– c.3435T>C polymorphism and breast cancer risk; (2) The total number of participants in the case and control groups, the genotype distribution, or other relevant information could be extracted from the paper. (3) Paper published in English language. The exclusion criteria for meta-analysis were as follow: (1) publication was review, meta-analysis, letter to the editor, or abstract; (2) the paper introduced either vague information or did not prepare frequencies of genotypes; (3) the paper related to ethnicities except than Asians.
The information from included papers were extracted by two authors independently (DK and MK). Differences concerning study selection and data extraction were settled by a discussion. We extracted the following information from eligible studies: first author’s name, publication year, country, genotyping methods, frequencies of genotypes in case and control groups, and estimation of Hardy-Weinberg equilibrium (HWE) in control groups.
At first, Hardy-Weinberg equilibrium (HWE) for control group of each study were calculated by a Chi-squared test. This calculation was performed by had2know online web server. A two-tailed p value less than 0.05 (p<0.05) was considered as a statistically significant difference. All statistical meta-analysis were done by Comprehensive Meta Analysis program (Biostat, Inc., Englewood, NJ, USA) and The Open Meta Analyst software (Tufts University, Medford, MA, USA). Pooled odds ratios (ORs) with 95% confidence intervals (CIs) were estimated to assess the association of MDR1– c.3435T>C polymorphism and breast cancer risk. Tests were performed under the following models: 1- Allelic (T vs. C allele), 2- Homozygous (TT vs. CC), 3- Heterozygous (CT vs. CC), and 4- Dominant model (TT+CT vs. CC), and 5- Recessive model (TT vs. CT+CC). The Cochrane Q-test and I2 score were employed to estimate heterogeneity and a p value more than 0.1 was considered as statistically significant. Fixed-effect and random-effects model were used in the absence and presence of heterogeneity of the studies, respectively. Also, a stratified meta-analysis was performed in Iranian populations. Publication bias was discovered by funnel plots and Egger’s test (Begg and Mazumdar, 1994; Egger et al., 1997). Sensitivity analysis was done to evaluate the magnitude effect of individual studies on the overall analysis.
In Silico Analysis
For bioinformatics analysis, the entire gene sequence of MDR1 was deduced from NCBI database (Chromosome 7, NC_000007.14). The location of c.3435T>C SNP on the MDR1 gene was determined manually. Also, the location of this SNP on RNA sequence was found according to the procedure. Since c.3435T>C mutation is a synonymous SNP, the effects of this transition on protein structure were not evaluated in the current study. But, this polymorphism may affect the RNA structure or/and splicing pattern of MDR1. Therefore, we used some bioinformatics tools to evaluate of these molecular effects of c.3435T>C on MDR1. For this purpose, RNAsnp server was used to evaluate the effects of c.3435T>C on the RNA structure (http://rth.dk/resources/rnasnp/).
The RNAsnp software works in three methods: Method 1 is planned to calculate the effect of SNPs on small RNA sequences (less than 1000 nucleotides); Method 2 is planned to expect the influence of SNPs on lengthy RNA sequence while the finally Method 3 works as a combination of method 1 and 2 and it is planned to screen all potential structure-wrecking single nucleotide polymorphisms in an input sequence using a brute-force methodology. In plot summary of the RNAsnp output, the local area which identified with maximum structural alteration is highlighted giving to the p-value. If the p-value is more than 0.2 (p> 0.2) the area is highlighted in black that shows not significant structural alteration is happened (Sabarinathan et al., 2013). The ASSP (http://wangcomputing.com/assp/) servers was employed to explore the impact of c.3435T>C on splice site pattern of MDR1. ASSP is a nucleotide sequence examination instrument for the estimation and cataloguing of splice sites. It works based on constitutive, cryptic, skipped, and alternative exon isoform splice sites analyses (Wang and Marín, 2006). ASSP recognizes putative splice sites by models which known as pre-processing models. Finally, we used String online web server to predict the gen-gene interaction network of MDR1.
Results
After screening of studies and as shown in figure 1, total of 7 eligible papers were included in the meta-analysis (Tatari et al. 2009; George et al. 2009 ; Taheri et al. 2010; Wu et al. 2012; Ghafouri et al. 2016; Abuhaliema et al., 2016; Sharif et al., 2016). The details of included studies in meta-analysis are given in table 1. This meta-analysis involved 1819 breast cancer patients and 1929 healthy controls. There were 4 studies with Iranian populations, and 3 remained studies are related to Indian, Chinese, and Jordanian populations. All of seven studies used polymerase chain reaction – restriction fragment length polymorphism (PCR-RFLP) method for c.3435T>C SNP genotyping. Also, the distribution of genotypes in all of control groups were consistent to Hardy–Weinberg criteria (Table 1).
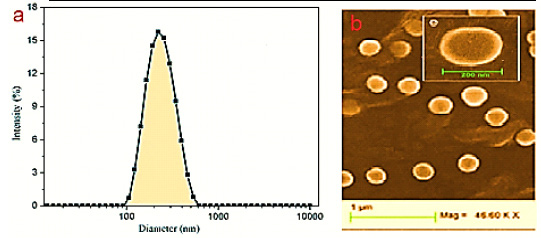 |
Figure 1: Study identification flowchart |
| Table 1: Characteristics of included studies in meta-analysis | |||||||||
| Country | Genotype frequencies | PHWE | Genotyping method | Author (Reference) | |||||
| Controls | Cases | ||||||||
| CC | CT | TT | CC | CT | TT | ||||
| Iran | 12 | 45 | 20 | 16 | 57 | 33 | 0.11 | PCR–RFLP | Tatari et al. 2009 |
| India | 15 | 32 | 21 | 8 | 39 | 39 | 0.67 | PCR–RFLP | George et al. 2009 |
| Iran | 10 | 27 | 13 | 10 | 30 | 14 | 0.55 | PCR–RFLP | Taheri et al. 2010 |
| China | 440 | 624 | 180 | 388 | 565 | 220 | 0.08 | PCR–RFLP | Wu et al. 2012 |
| Iran | 141 | 50 | 9 | 75 | 16 | 9 | 0.11 | PCR–RFLP | Ghafouri et al. 2016 |
| Jordan | 40 | 65 | 45 | 68 | 62 | 20 | 0.10 | PCR–RFLP | Abuhaliema et al., 2016 |
| Iran | 79 | 53 | 8 | 61 | 68 | 21 | 0.82 | PCR-RFLP | Sharif et al., 2016 |
| HWE: Hardy–Weinberg equilibrium, PCR-RFLP: Polymerase chain reaction-restriction fragment length polymorphism. A Hardy–Weinberg equilibrium in the control group with P <0.05 did not satisfy the Hardy-Weinberg equilibrium. | |||||||||
Results of association between mentioned SNP and risk of breast cancer are summarized in table 2. Our data revealed that there are no significant associations between MDR1 c.3435T>C and risk of breast cancer in Asian population under all of five genetic models (T vs. C: OR= 1.22, 95%CI= 0.86-1.75, P= 0.267; TT vs. CC: OR= 1.35, 95%CI= 0.72-2.55, P= 0.353; CT vs. CC: OR= 1.00; 95%CI= 0.72-1.38; P= 0.993; CT+TT vs. CC: OR= 1.07, 95%CI= 0.73-1.56, P= 0.724; TT vs. CC+CT: OR= 1.26, 95%CI= 0.80-1.99, P= 0.323) (Figure 2). When we performed a stratified meta-analysis in Iranian populations, we found a significant association between MDR1 c.3435T>C and breast cancer risk in Iranian populations under three genetic models (T vs. C: OR= 1.40, 95%CI= 1.14-1.73, P= 0.002; TT vs. CC: OR= 1.85, 95%CI= 1.15-2.98, P= 0.011; TT vs. CC+CT: OR= 1.59, 95%CI= 1.06-2.37, P= 0.024) (Figures 3 and 4).
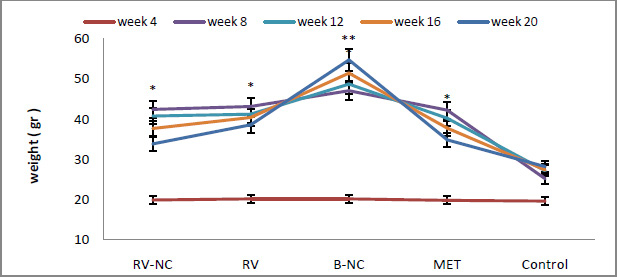 |
Figure 2: Forest plot for Asian meta-analysis. A) CT vs. CC model; B) CT+TT vs. CC model; C) TT vs. CC+CT model; D) TT vs. CC model. |
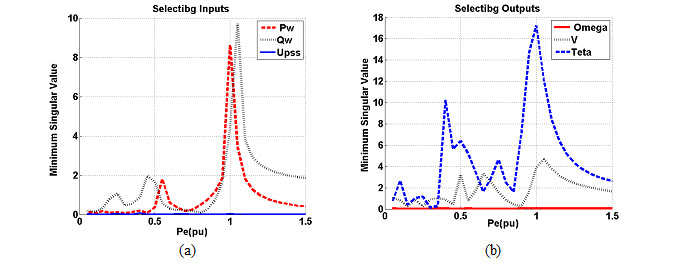 |
Figure 3 |
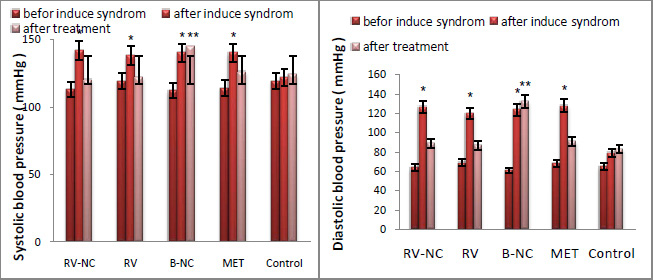 |
Figure 4: Forest plot for Iranian populations. A) CT vs. CC model; B) CT+TT vs. CC model. |
The results of heterogeneity and publication bias are detailed in table 3. Our data revealed that there are true heterogeneities in overall analysis under all of five genetic models (T vs. C: Pheterogeneity < 0.001; I2=88%; TT vs. CC: Pheterogeneity< 0.001; I2=81%; CT vs. CC: Pheterogeneity= 0.028; I2=58%; CT+TT vs. CC: Pheterogeneity< 0.001; I2=75%; TT vs. CC+CT: Pheterogeneity< 0.001; I2=75%). Whereas we observed only a significant heterogeneity in stratified analysis for Iranian population under CT vs. CC model (Pheterogeneity= 0.095; I2=53%). Publication bias was determined by Egger’s test and funnel plot. As shown in table 3 there are no publication biases in this meta-analysis in overall analysis (T vs. C: PEgger= 0.780; TT vs. CC: PEgger= 0.944; CT vs. CC: PEgger= 0.989; CT+TT vs. CC: PEgger= 0.991; TT vs. CC+CT: PEgger= 0.947) and stratified analysis (T vs. C: PEgger= 0.125; TT vs. CC: PEgger= 0.296; CT vs. CC: PEgger= 0.600; CT+TT vs. CC: PEgger= 0.592; TT vs. CC+CT: PEgger= 0.604). This data were confirmed by symmetrical funnel plots (Figure 5). To assess the strength of the association results, sensitivity analysis was done by eliminating one study at a time and recalculating the summary ORs. The summary ORs continued stable, representing that our meta-analysis is not significantly influenced by an individual study (data not shown).
 |
Figure 5: Funnel plot for Asian meta-analysis. A) CT vs. CC model; B) TT vs. CC model; C) CT+TT vs. CC model; D) TT vs. CC+CT model. |
The data from ASSP online web server showed that the c.3435T>C polymorphism alters splice site pattern of the MDR1 gene (Table 4). The data revealed that the score of constitutive donor splice site at location 515 is 6.381 for 3435TT genotype whereas this score for 3435CC genotype is 5.066. In addition Alt./Cryptic, constitutive, and confidence scores for genotype at location 507 are 0.652, 0.334, and 0.487, respectively. Whereas these values for 3435CC are 0.628, 0.335, and 0.434, respectively. In addition, all of mentioned values are different between 3435TT and 3435CC genotypes at location 515 (0.914, 0.057, and 0.937 for TT genotype and 0.820, 0.136, and 0.834 for CC genotype).
To evaluate the effects of c.3435T>C SNP on mRNA structure of MDR1, we used the RNAsnp server. For this purpose, we entered the entire coding sequence of the gene into RNAsnp main window. Then we entered the code of SNP in a section of server entitled “SNP details”. The data from RNAsnp revealed that c.3435T>C could not significantly affect RNA structure of MDR1 gene (Folding Window=3235-3635, Local region=3434-3483, distance=0.0965, p-value=0.2983). Also, we found that the minimum free energy of the optimal secondary structure of global TT genotype sequence is -113.70 kcal/mol, while this is -114.60 kcal/mol for CC genotype.
| Table 2: Association results of MDR1 c.3435T>C and breast cancer risk in the meta-analysis | ||||||||||
| Group | T vs. C | TT vs. CC | CT vs. CC | CT+TT vs. CC | TT vs. CC+CT | |||||
| OR
(95% CI) |
P | OR
(95% CI) |
P | OR
(95% CI) |
P | OR
(95% CI) |
P | OR
(95% CI) |
P | |
| Asian | 1.22
(0.86-1.75) |
0.267 | 1.35
(0.72-2.55) |
0.353 | 1.00
(0.72-1.38) |
0.993 | 1.07
(0.73-1.56) |
0.724 | 1.26
(0.80-1.99) |
0.323 |
| Iranian | 1.40
(1.14-1.73) |
0.002 | 1.85
(1.15-2.98) |
0.011 | 1.04
(0.62-1.74) |
0.895 | 1.24
(0.92-1.68) |
0.160 | 1.59
(1.06-2.37) |
0.024 |
| OR: odds ratio, CI: confidence interval | ||||||||||
Discussion
Breast cancer is known as the most common malignancy among women in both developing and developed countries. This disorder is a rising issue that influences about 12.5% of women during their life. The rate of this malignancy is on the growth (Slamon et al., 1987; Soleimani et al., 2016). The occurrence of breast cancer in Asian women is in overall lower than other ethnicity. But, all health statistics showed that this malignancy has been quickly growing in current decades in Asia (Matsuno et al., 2007).
The reasons of breast cancer are weakly recognized. Some factors including family history, age, lifestyle, geographical variation, and genetic factors may increase the risk of breast cancer. Genetic variations are widely considered as important risk factors for breast cancer susceptibility in exact populations (Haimov-Kochman et al., 2002). Recognition of some crucial single nucleotide polymorphisms, which impacts the gene expression and protein function, it might be suitable to predict susceptibility of breast cancer (Lian et al., 2012).
| Table 3: Results of heterogeneity and publication bias for the association of MDR1 c.3435T>C and breast cancer risk | |||||||||||||||
| Group | T vs. C | TT vs. CC | CT vs. CC | CT+TT vs. CC | TT vs. CC+CT | ||||||||||
| Ph | I2 | Pe | Ph | I2 | Pe | Ph | I2 | Pe | Ph | I2 | Pe | Ph | I2 | Pe | |
| Asian | < 0.001 | 88% | 0.780 | < 0.001 | 81% | 0.944 | 0.028 | 58% | 0.989 | < 0.001 | 75% | 0.991 | < 0.001 | 75% | 0.947 |
| Iranian | 0.140 | 45% | 0.125 | 0.335 | 12% | 0.296 | 0.095 | 53% | 0.600 | 0.117 | 49% | 0.592 | 0.351 | 8% | 0.604 |
| Ph: Pheterogeneity (p< 0.1) was considered as a significant difference. Pe: PEgger (p< 0.05) was considered as a significant difference | |||||||||||||||
In this study, we investigate the association of MDR1 c.3435T>C polymorphism and breast cancer risk in Asian population by a meta-analysis which followed by a bioinformatics approach. For example Tatari et al. (2009) reported a significant association between MDR1-3435T allele and risk of breast cancer, whereas Taheri et al. (2010) didn’t discover any association. Therefor meta-analysis as an influential statistical instrument can help researchers to find out more precise conclusions. Our meta-analysis revealed that there are no significant association between mentioned SNP and breast cancer risk in overall Asian analysis. But, stratified meta-analysis in Iranian population showed a significant association between MDR1 c.3435T>C transition and risk of breast cancer in some genetic model. Also, after stratifying analysis we found that the heterogeneity become disappear. Therefore, it may be a source of heterogeneity among the studies. The different results in various studies may be due to environmental factors, genetic backgrounds and lifestyles.
| Table 4: ASSP prediction results for 3435TT and 3435CC genotypes | |||||||
| Positio (bp) | Putative splice site | Sequence | Score | Intron GC | Activations | Confidence | |
| Alt./Cryptic | Constitutive | ||||||
| A) 3435TT Genotype | |||||||
| 458 | Alt. isoform/cryptic acceptor | ttgactgcagCATTGCTGAG | 6.509 | 0.586 | 0.696 | 0.300 | 0.569 |
| 507 | Alt. isoform/cryptic acceptor | ggtgtcacagGAAGAGATTG | 2.614 | 0.543 | 0.652 | 0.334 | 0.487 |
| 515 | Alt. isoform/cryptic donor | GGAAGAGATTgtgagggcag | 6.381 | 0.486 | 0.914 | 0.057 | 0.937 |
| 569 | Constitutive donor | ACTGCCTAATgtaagtctct | 13.099 | 0.429 | 0.103 | 0.852 | 0.879 |
| B) 3435CC Genotype | |||||||
| 458 | Alt. isoform/cryptic acceptor | ttgactgcagCATTGCTGAG | 6.509 | 0.586 | 0.696 | 0.300 | 0.569 |
| 507 | Alt. isoform/cryptic acceptor | ggtgtcacagGAAGAGATCG | 2.614 | 0.543 | 0.628 | 0.355 | 0.434 |
| 515 | Alt. isoform/cryptic donor | GGAAGAGATCgtgagggcag | 5.066 | 0.486 | 0.820 | 0.136 | 0.834 |
| 569 | Constitutive donor | ACTGCCTAATgtaagtctct | 13.099 | 0.429 | 0.103 | 0.852 | 0.879 |
Some mechanisms explain the role of MDR1 gene in breast cancer risk. MDR1 gene also known as MDR1 is a member of the MDR1 superfamily that expresses P-gp protein, which is an ATP-dependent efflux pump that permits the human cells to remove poisons and carcinogenic agents (Kreile et al., 2013). Some studies proposed that c.3435T>C transition may impact the risk of some cancers such as breast carcinoma (Ikeda et al., 2015). Really, this synonymous transition (Ile1145Ile) effects stability of protein (Fung et al., 2014) and leads to apoptosis modification or cellular damage witch these are important for development of cancer (Johnstone et al., 2000).
Our previous in silico studies revealed that SNPs may affect gene expression (Jamali et al., 2016) mRNA structure (Karimian et al., 2015; Raygan et al., 2016) and protein structure and function (Karimian and Hosseinzadeh Colagar, 2016a; Nikzad et al., 2015; Karimian and Hosseinzadeh Colagar, 2016b).
The SNP which was studied in this study is a synonymous polymorphism, and then it has no effect on the peptide sequence of the protein. But, it could impact the mRNA structure or splicing pattern. So it is possible to apply bioinformatics tools to assess the harmful properties of MDR1– c.3435T>C polymorphism on the splicing pattern and mRNA structure. So that we used in silico tools to evaluate these effects. Our data from in silico analysis revealed that c.3435T>C could not affect the mRNA structure. Though c.3435T>C substitution leads to a decrease in minimum free energy for mutant type but this change is not significant. Therefore, it is expected that pathogenicity of c.3435T>C may arise from its effect on RNA splicing.
There are some limitations in our meta-analysis that should be mentioned. For instance, lack of original data, such as smoking, BMI, age, and etc. may affect the accuracy of the association of the SNP and risk of breast cancer. Also, this meta-analysis did not cover a large number of Asian countries. In conclusion, the present study suggests that the MDR1– c.3435T>C transition might be correlated with risk breast cancer in Iranian population. But, studies with larger sample size and considered to gene-environment and gene-gene interactions are essential to approve our results.
References
Abuhaliema, A.M., Yousef, A.M., El-Madany, N.N., Bulatova, N.R., Awwad, N.M., Yousef, M.A., & Al Majdalawi, K.Z. (2016) Influence of Genotype and Haplotype of MDR1 (C3435T, G2677A/T, C1236T) on the Incidence of Breast Cancer-a Case-Control Study in Jordan. Asian Pacific Journal of Cancer Prevention 17(1), 261-266.
Egger, M., Davey Smith, G., Schneider, M., Minder, C. (1997) Bias in metaanalysis detected by a simple, graphical test. BMJ 315, 629-634.
Fung, K. L., Pan, J., Ohnuma, S., Lund, P. E., Pixley, J. N., Kimchi-Sarfaty, C.& Gottesman, M. M. (2014) MDR1 synonymous polymorphisms alter transporter specificity and protein stability in a stable epithelial monolayer. Cancer Research 74(2), 598-608.
George, J., Dharanipragada, K., Krishnamachari, S., Chandrasekaran, A., Sam, S. S., & Sunder, E. (2009) A single-nucleotide polymorphism in the MDR1 gene as a predictor of response to neoadjuvant chemotherapy in breast cancer. Clinical Breast Cancer 9(3), 161-165.
Ghafouri, H., Ghaderi, B., Amini, S., Nikkhoo, B., Abdi, M., & Hoseini, A. (2016) Association of ABCB1 and ABCG2 single nucleotide polymorphisms with clinical findings and response to chemotherapy treatments in Kurdish patients with breast cancer. Tumor Biology 37(6), 7901-7906.
Haimov-Kochman, R., Lavy, Y., Hochner-Celinkier, D. (2002) Review of risk factors for breast cancer-what’s new? Harefuah 141, 702-708.
Ikeda, M., Tsuji, D., Yamamoto, K., Kim, Y. I., Daimon, T., Iwabe, Y. & Nakamichi, H. (2015) Relationship between ABCB1 gene polymorphisms and severe neutropenia in patients with breast cancer treated with doxorubicin/cyclophosphamide chemotherapy. Drug Metabolism and Pharmacokinetics, 30(2), 149-153.
Isvoran, A., Louet, M., Vladoiu, D. L., Craciun, D., Loriot, M. A., Villoutreix, B. O., & Miteva, M. A. (2016) Pharmacogenomics of the cytochrome P450 2C family: impacts of amino acid variations on drug metabolism. Drug Discovery Today. Doi: 10.1016/j.drudis.2016.09.015.
Jamali, S., Karimian, M., Nikzad, H., & Aftabi, Y. (2016) The c.− 190 C> A transversion in promoter region of protamine1 gene as a genetic risk factor for idiopathic oligozoospermia. Molecular Biology Reports 43, 795-802.
Jemal, A., Siegel, R., Ward, E., Murray, T., Xu, J., Smigal, C., & Thun, M. J. (2006) Cancer statistics, 2006. CA: a Cancer Journal for Clinicians, 56(2), 106-130.
Johnatty, S. E., Beesley, J., Gao, B., Chen, X., Lu, Y., Law, M. H. & Fereday, S. (2013) ABCB1 (MDR1) polymorphisms and ovarian cancer progression and survival: a comprehensive analysis from the Ovarian Cancer Association Consortium and The Cancer Genome Atlas. Gynecologic Oncology, 131(1), 8-14.
Johnstone, R. W., Ruefli, A. A., & Smyth, M. J. (2000) Multiple physiological functions for multidrug transporter P-glycoprotein? Trends in Biochemical Sciences, 25(1), 1-6.
Karimian, M., Hosseinzadeh Colagar, A. (2016a) Methionine synthase A2756G transition might be a risk factor for male infertility: evidences from seven case-control studies. Molecular and Cellular Endocrinology 425, 1–10.
Karimian, M., Hosseinzadeh Colagar, A. (2016b) Association of C677T transition of the human methylenetetrahydrofolate reductase (MTHFR) gene with male infertility. Reproduction Fertility and Development 28, 785-794.
Karimian, M., Nikzad, H., Azami-Tameh, A., Taherian, A., Darvishi, F. Z., & Haghighatnia, M. J. (2015) SPO11-C631T gene polymorphism: association with male infertility and an in silico-analysis. Journal of Family and Reproductive Health 9, 155-163.
Kreile, M., Rots, D., Piekuse, L., Cebura, E., Grutupa, M., Kovalova, Z., & Lace, B. (2013) Lack of association between polymorphisms in genes MTHFR and MDR1 with risk of childhood acute lymphoblastic leukemia. Asian Pacific Journal of Cancer Prevention: APJCP, 15(22), 9707-9711.
Lee, I. H. (2016) Molecular Prognostic and Predictive Assays in Breast Cancer: A Practical Review. Pathology Case Reviews, 21(1), 4-10.
Lian, H., Wang, L., Zhang, J. (2012) Increased risk of breast cancer associated with CC genotype of Has-miR-146a Rs2910164 polymorphism in Europeans. PLoS One 7, e31615.
Matsuno, R. K., Anderson, W. F., Yamamoto, S., Tsukuma, H., Pfeiffer, R. M., Kobayashi, K., … & Levine, P. H. (2007) Early-and late-onset breast cancer types among women in the United States and Japan. Cancer Epidemiology Biomarkers & Prevention, 16(7), 1437-1442.
Nickels, S., Truong, T., Hein, R., Stevens, K., Buck, K., Behrens, S. & Gaudet, M. (2013) Evidence of gene-environment interactions between common breast cancer susceptibility loci and established environmental risk factors. PLoS Genet, 9(3), e1003284.
Nikzad, H., Karimian, M., Sareban, K., Khoshsokhan, M., & Colagar, A. H. (2015) MTHFR-Ala222Val and male infertility: a study in Iranian men, an updated meta-analysis and an in silico analysis. Reproductive Biomedicine Online 31, 668-680.
Ozdemir, S., Uludag, A., Silan, F., Atik, S. Y., Turgut, B., & Ozdemir, O. (2013) Possible roles of the xenobiotic transporter P-glycoproteins encoded by the MDR1 3435 C> T gene polymorphism in differentiated thyroid cancers. Asian Pacific Journal of Cancer Prevention, 14(5), 3213-3217.
Raygan, F., Karimian, M., Rezaeian, A., Bahmani, B., & Behjati, M. (2016) Angiotensinogen-M235T as a risk factor for myocardial infarction in Asian populations: a genetic association study and a bioinformatics approach. Croatian Medical Journal 57, 351-362.
Sabarinathan, R., Tafer, H., Seemann, S. E., Hofacker, I. L., Stadler, P. F., & Gorodkin, J. (2013) The RNAsnp web server: predicting SNP effects on local RNA secondary structure. Nucleic Acids Research 41(W1), W475-W479.
Sharif, A., Kheirkhah, D., Sharif M.R., Karimian M., Karimian, Z. (2016) ABCB1-C3435T polymorphism and breast cancer risk: a case-control study and a meta-analysis. JBUON, 21(6), 1433-1441.
Slamon, D. J., Clark, G. M., Wong, S. G., Levin, W. J., Ullrich, A., & McGuire, W. L. (1987) Human breast cancer: correlation of relapse and. Science, 3798106(177), 235.
Soleimani, Z., Kheirkhah, D., Sharif, M. R., Sharif, A., Karimian, M., & Aftabi, Y. (2016) Association of CCND1 Gene c. 870G> A Polymorphism with Breast Cancer Risk: A Case-ControlStudy and a Meta-Analysis. Pathology & Oncology Research, 1-11.
Taheri, M., Mahjoubi, F., Omranipour, R. (2010) Effect of MDR1 polymorphism on multidrug resistance expression in breast cancer patients. Genetics and Molecular Research 9, 34-40.
Tatari, F., Salek, R., Mosaffa, F., Khedri, A., & Behravan, J. (2009) Association of C3435T single-nucleotide polymorphism of MDR1 gene with breast cancer in an Iranian population. DNA and Cell Biology, 28(5), 259-263.
Tazzite, A., Kassogue, Y., Diakité, B., Jouhadi, H., Dehbi, H., Benider, A., & Nadifi, S. (2016) Association between ABCB1 C3435T polymorphism and breast cancer risk: a Moroccan case-control study and meta-analysis. BMC Genetics, 17(1), 126.
Turgut, S., Yaren, A., Kursunluoglu, R., et al. (2007) MDR1 C3435T polymorphism in patients with breast carcinoma. Archives of Medical Research 38, 539-544.
Wang, M., & Marín, A. (2006) Characterization and prediction of alternative splice sites. Gene 366(2), 219-227.
Wu, H., Kang, H., Liu, Y., Tong, W., Liu, D., Yang, X. & Sha, X. (2012) Roles of ABCB1 gene polymorphisms and haplotype in susceptibility to breast carcinoma risk and clinical outcomes. Journal of Cancer Research and Clinical Oncology, 138(9), 1449-1462.


