1Harcourt Butler Technical University, Kanpur
2Naraina Vidyapeeth Engineering & Management Institute, Kanpur
Corresponding author Email: drvinay@yahoo.com
Article Publishing History
Received: 12/07/2019
Accepted After Revision: 18/09/2019
Lipase or triacylglycerol acyl ester hydrolases belong to serine hydrolase family also called as carboxylic acid esterases, whose number is EC 3.1.1.3 according to enzyme-commission that helps to break the bond by reaction with water. Microbial lipase production is preferable than plants and animals because of the more rapid growth of microbes, ease to genetic manipulation, requires low-cost media, stability and their specific properties. Currently, microbial lipase has many applications in the industrial field. Agro-industrial by-products are used in the biotechnology field because it contains carbon, nitrogen, minerals and other nutrients, and they are of low cost. Production of lipase by micro-organism is carried out with the help of agriculture by-product using them as substrate. In mix culture experiment in case of each substrate, maximum lipase activity was obtained using mustard oil cake (10.199458 U/ml), sesame oil cake (10.6731 U/ml), linseed oil cake (9.947174941 U/ml), and soybean oil cake (9.5716 U/ml). Overall mix culture experiment, sesame oil cake gave highest lipase activity.
Agriculture Residues; Mustard Oil Cake, Linseed Oil Cake, Sesame Oil Cake, and Soybean Oil Cake; Optimization; Lipase; Mix Culture; Submerged Fermentation
Prabha S, Verma G, Pandey S, Singh. B, Dwivedi V. Utilization of Agro-Industrial By-Products for Production of Lipase Using Mix Culture Batch Process. Biosc.Biotech.Res.Comm. 2019;12(3).
Prabha S, Verma G, Pandey S, Singh. B, Dwivedi V. Utilization of Agro-Industrial By-Products for Production of Lipase Using Mix Culture Batch Process. Biosc.Biotech.Res.Comm. 2019;12(3). Available from: https://bit.ly/2kzpMPT
Copyright © Prabha et al., This is an open access article distributed under the terms of the Creative Commons Attribution License (CC-BY) https://creativecommns.org/licenses/by/4.0/, which permits unrestricted use distribution and reproduction in any medium, provide the original author and source are credited.
Introduction
Lipase or triacylglycerol acyl ester hydrolases belong to serine hydrolase family also called as carboxylic acid esterases, whose number is EC 3.1.1.3 according to enzyme-commission that helps to break the bond by reaction with water. Protease, amidase, glucosidase, nitrilases, epoxide enzyme also belongs to a hydrolase family (Patrick Fickers et. al., 2011) (R. Gupta et. al., 2004); (Hasan et al., 2006); (Hasan et. al., 2009); (Jaeger & Eggert, 2002); (Salihu et. al., 2012). Conventionally, lipase enzyme was obtained from animal sources. Lipase enzyme was firstly discovered by Claud Bernard in 1856 in pancreatic juice, which converts the insoluble oil-fats to the soluble product (F. Hasan et al., 2006). Pancreatic lipase is present with many other enzymes such as trypsin, which gives bitter taste with undesirable effect. Lipase enzyme easily extracted from plants but the process in fermenter performed is complicated and increase the production cost. So plant lipase has not used for commercial application. Microbial lipase production is preferable than plants and animals because of the more rapid growth of microbes, ease to genetic manipulation, requires low-cost media, stability and their specific properties (Rocha, Padez, & Morais, 1998). First lipase production was carried out by Bacillus prodigiosus, Bacillus pyocyaneus, and Bacillus fluorescent in 1901. Maximum lipase production by the microorganism is based on different strains and characteristics, specificity, stability, performance, and mechanism of action (Bornscheuer et. al., 2013) (Sharma et. al., 2011,Geoffry and Archer 2018).
So the problems of lipase production by animals and plants overcome by microbial lipase. Microbial lipase attracted more attention due to low production cost and has economic importance. Now a day, the demand for lipase is fulfilled from micro-organism like bacteria, yeast, fungi, and actinomycetes, which produce vast diversity of extracellular lipase (Sharma et al., 2011). Fermentation process for the lipase production by SmF and SSF in the last few years. Submerged fermentation and solid state fermentation technique are the most common and conventional process used for lipase production which has an advantage over the other processes ( Sun & Xu, 2008). Agro-industrial by-products are used in the biotechnology field because it contains carbon, nitrogen, minerals and other nutrients, and they are of low cost. Production of lipase by micro-organism is carried out with the help of agriculture by-product using them as substrate. Waste as substrate contains a high amount of nutrients and minerals that are essential for the growth of microorganism. Sometimes substrates act as an inducer for microbial growth. The substrate that has an essential lipid component required for the production of lipase is called an ideal substrate. Otherwise, assemble essential component then provides the value-added supplements for the growth of microorganism (Pandey et al., 1999) (Mitchell et. al., 2000) (Martínez-Herrera et. al., 2006). Much agriculture residual are converted to renewable product for using as substrate for lipase production using microorganism. Lipids are present in oil-cake after extraction of oil in industries(Singhania et. al., 2008 Hasan et al 2018).
Currently, microbial lipase has many applications in the industrial field. Esterification and transesterification reaction demand is increasing day by day because the lipase enzyme has performed the mechanism in non-aqueous media condition. (Mendes et. al., 2012) (F. Hasan et al., 2006) (Sun et. al., 2013) (Kapoor & Gupta, 2012). Microbial lipase has broad application in another biotechnological field such as leather, textile, pulp and paper, cosmetics, and fat- oil industries.
Table 1: Lipase producing microorganisms with the production process and lipase activity
| S. No. | Micro-organism | Lipase activity | References |
| 1. | Yarrowia lipolytica YlLip2 | 42900 | (Yu, Wen, & Tan, 2010) |
| 2. | Candida cylindracea CBS786,
Candida rugosa CBS2275, Yerrowia lipolytica W29 (ATCC20460) |
30 U/L/h
20 U/L/h 7 U/L/h |
(Gonçalves, Oliveira et. al., 2012) |
| 3. | Pseudomonas aeruginosa | 204.12 U/mg | (Zouaoui & Bouziane, 2012) |
| 5. | Bacteria SSB1N | 0.1128 µg/ml/min | (N. A. Hasan et. al., 2018) |
| 6. | Serratia marcescens ECU1010 | 640 U/g | (Long, Xu, & Pan, 2007) |
| 7. | Garbage lipase enzyme | 57.43 U/ml | (Selvakumar & Sivashanmugam, 2017) |
| 8. | Pseudomonas aeruginosa | 60 U/ml | (Saravanan et. al., 2007) |
| S. No. | Micro-organism | Lipase Activity | References |
| 9. | Fusarium solani NFCCI 4084 | 7.8 U/ml | (Geoffry & Achur, 2018) |
| 10.. | Thermomyces
lanuginosus (GSLMBKU-10, GSLMBKU-13, GSLMBKU-14) |
205.80 µg/ml, 225.30 µg/ml, 165.23 µg/ml | (Sreelatha et. al., 2017) |
| 11. | Aspergillus niger J-1 | 1.46 IU/ml
In SmF, 4.8 IU/ml in SSF |
(Falony et. al., 2006) |
| 12. | Aspergillus niger AS-02 | 49.37 U/g | (Salihu et. al., 2016) |
| 13. | Yarrowia lipolytica (CECT 1240) | 57.9 U/cm³ | (Domínguez et. al.,2003) |
| 14. | Candida cylindracea
( NRRL Y-17506) |
9.231 IU/ml | (D’Annibale et. al., 2006a) |
| 15. | Bacillus licheniformis 016 | 1870 U/L | (Baltaci et. al., 2018) |
| 16. | E.coli BL21 (DE3) | 206 U/ml | (Chai et. al., 2018) |
| 17. | Penicillium simplicissimum | 44.8 U/g | (Godoy et al., 2009) |
| 18. | Penicillium simplicissimum | 30 Ugd/s | (Asenjo & Andrews, 2008) |
| 20. | Bacillus subtilis | 4.96 U/ml | (Suci et. al., 2018) |
| 21. | Penicillum restrictum | 30.3 U/g | (Gombert, Pinto et. al., 1999) |
| 22. | Aspergillus oryzae NCIM 1212,
Aspergillus japonicas MTCC 1975 |
18.9 U/g
23 U/g |
(Jain & Naik, 2018) |
| 23. | Bacillus stratosphericus PSP8 | 47 U/ml | (Ismail et al., 2018) |
| 24. | Yarrowia lipolytica | 68.03 U/g | (da S. Pereira et. al., 2019). |
| 25. | Bacillus coagulans | 78069 U/g | (Alkan et. al., 2007) |
| 26. | Rhodotorula glutinis HL25 | 75.2 U/l | (Taskin et al., 2016) |
| 27. | Penicillium gracilenta CBMAI 1583 | 1.62 U/ml | (Turati et al., 2019) |
| 28. | Bacillus cereus | 117.3±20 U/ml | (Vasiee et. al., 2016) |
| 29. | Penicillium P58 and P74 | 139.2 U lipase/g
140.7 U lipase/g |
(Rigo et al., 2010) |
Materials and Methods
Culture procuration
Bacillus licheniformis MTCC 3244 and Bacillus coagulans MTCC 10305 was procured from Microbial Culture Collection and gene bank (MTCC), Institute of Microbial Technology, Chandigarh, India. Both stock cultures were maintained on media composition containing (g/l) Yeast extract, 2; Beef extract, 1; Peptone, 5; NaCl, 5; Agar, 15.
Substrates collection & pretreatment
Mustard oil cake, linseed oil cake, sesame oil cake and soybean oil cake were collected from the local market at Kalyanpur, Kanpur (U.P.). Cakes were dried in a hot air oven at 75℃ for 2 to 3 days and were ground in a mixer. Cakes were stained by strainer (0.5mm) to obtain fine powder form. The powder form of cakes was stored in airtight containers. Powder form cakes were then used as substrate.
Lipase production by submerged fermentation
Lipase production using selected strain Bacillus lichenifomis MTCC3244 and Bacillus coagulans 10305 was carried out by submerged fermentation. A semi-synthetic liquid medium containing (%) Glucose, 1; Peptone, 1; Yeast extract, 0.5; MgSO4.7H2O, 0.05; KCl, 0.05; FeSO4.7H2O, 0.001; Olive oil, 0.3%; Substrate (cake oil), 2.5 (Gutarra et al.,2009) was adjust the different pH then sterilized by autoclaving at 121℃,15 psi for 15 min. Flasks were inoculated with of 0.25% Bacillus lichenifomis MTCC3244 and Bacillus coagulans 10305 was added 20 ml of production medium (in Erlenmeyer flask of 100 ml volume). The Erlenmeyer flasks were incubated at different pH, temperature (℃) and inoculum concentration (%) on rotary shaker at 100 rpm. Each 1 ml samples were then centrifuged for 5 min at 12000 rpm. The supernatant was collected. The absorbance of the culture was measured by spectrophotometer at wavelength 410 nm. Maximum lipase activity was achieved by different parameters pH (4, 5, 6, 7, 8, 9), temperature (35℃, 40℃, 45℃, 50℃) and inoculum concentration (5%, 10%, 15%). Medium optimization studies were carried out by studying one-factor-at-a-time and all factors were kept same.
Lipase activity determination
The activity of extracellular lipase was measured by an assay to measure the amount of para-nitophenol (PNP) formed from p-nitrophenol acetate (pNPA). In the assay, 2.87 ml of 100 mM potassium phosphate buffer (KPB) at pH 7 was added to the 100 µl culture supernatant in the test tube. After preincubation at 30℃ for 3 minute, the reaction was started by quick mixing the solution with 30 µl of 100 mM p-NPA solution in DMSO. After 10 minute, change in absorbance at 410 nm was recorded with a spectrophotometer. Amount of PNP formed was calculated by using standard prepared at different dilutions of PNP. One lipase unit was defined as amount of lipase enzyme required to convert 1µmole of pNPA to PNP per minute under above condition (Long, Xu, & Pan, 2007).
Results and Discussion
Culture procuration
 |
Figure 1: Growth of Bacillus licheniformis MTCC 3244 and Bacillus coagulans MTCC 10305 |
Optimization of process parameter by using the one-factor-at-a-time (OFAT) method
pH
Lipase activity of mix culture at different pH using mustard oil cake as substrate
As shown in table 2, maximum lipase activity was observed at pH 8 that was 9.9269 U/ml after 48 hr.
Table 2: Lipase activity of mix culture at different pH using mustard oil cake as substrate
| pH | OD at different time interval (410nm) | |||
| 24 Hr | Activity of lipase (U/ml) | 48Hr | Activity of lipase (U/ml) | |
| 4 | 0.449 | 2.3118 | 0.321 | 1.65275 |
| 5 | 0.623 | 3.2077 | 0.354 | 1.8227 |
| 6 | 0.899 | 4.62875 | 0.867 | 4.464 |
| 7 | 1.063 | 5.473 | 0.661 | 3.40335 |
| 8 | 1.807 | 9.304 | 1.928 | 9.9269 |
| 9 | 1.562 | 8.04245 | 1.632 | 8.40285 |
Lipase activity of mix culture at different pH using linseed oil cake as substrate
As shown in table 3, Maximum lipase activity was observed at pH 8 that was 9.74155 U/ml after 48 hr.
Table 3: Lipase activity of mix culture at different pH using linseed oil cake as substrate
| pH | OD at different time interval (410nm) | |||
| 24 Hr | Activity of lipase (U/ml) | 48Hr | Activity of lipase (U/ml) | |
| 4 | 0.612 | 3.15105 | 0.773 | 3.98 |
| 5 | 0.66 | 3.3982 | 0.801 | 4.1242 |
| 6 | 0.673 | 3.46515 | 0.837 | 4.30955 |
| 7 | 1.362 | 7.01265 | 1.342 | 6.9097 |
| 8 | 1.617 | 8.32535 | 1.892 | 9.74155 |
| 9 | 1.492 | 7.682 | 1.532 | 7.88795 |
Lipase activity of Mix culture at different pH using sesame oil cake as substrate
As shown in table 4, Maximum lipase activity was observed at pH 8 that was 10.3646 U/ml after 24 hr.
Table 4: Lipase activity of Mix culture at different pH using sesame oil cake as substrate
| pH | OD at different time interval (410nm) | |||
| 24 Hr | Activity of lipase (U/ml) | 48Hr | Activity of lipase (U/ml) | |
| 4 | 0.484 | 2.492 | 0.632 | 3.25405 |
| 5 | 0.205 | 1.0555 | 0.493 | 2.53835 |
| 6 | 0.684 | 3.5218 | 1.324 | 6.817 |
| 7 | 1.857 | 9.5613 | 1.699 | 8.7478 |
| 8 | 2.013 | 10.3646 | 1.681 | 8.65515 |
| 9 | 1.108 | 5.70485 | 0.521 | 2.6825 |
Lipase activity of Mix culture at different pH using soybean oil cake as substrate
As shown in table 5, Maximum lipase activity was observed at pH 8 that was 9.5716 U/ml after 24 hr.
| pH | OD at different time interval (410nm) | |||
| 24 Hr | Activity of lipase (U/ml) | 48Hr | Activity of lipase (U/ml) | |
| 4 | 0.484 | 2.492 | 0.213 | 1.0967 |
| 5 | 0.708 | 3.64535 | 0.504 | 2.595 |
| 6 | 0.728 | 3.74835 | 0.739 | 3.80495 |
| 7 | 0.968 | 4.98405 | 1.312 | 6.75525 |
| 8 | 1.859 | 9.5716 | 1.727 | 8.892 |
| 9 | 1.521 | 7.83135 | 1.632 | 8.40285 |
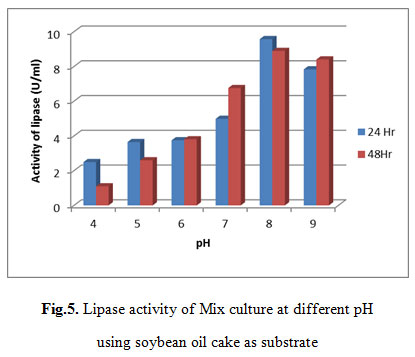 |
Figure 5: Lipase activity of Mix culture at different pH using soybean oil cake as substrate |
Temperature
Lipase activity of mix culture at different temperature using mustard oil cake as substrate
As shown in table 6, Maximum lipase activity was observed at 40℃ that was 6.73465 U/ml after 24 hr.
Table 6: Lipase activity of mix culture at different temperature using mustard oil cake as substrate
| Temp. (℃) | OD at different time interval (410nm) | |||
| 24 Hr | Activity of lipase (U/ml) | 48Hr | Activity of lipase (U/ml) | |
| 35 | 1.171 | 6.02925 | 1.095 | 5.63795 |
| 40 | 1.308 | 6.73465 | 1.265 | 6.51325 |
| 45 | 1.155 | 5.94685 | 1.065 | 5.48345 |
| 50 | 0.408 | 2.1007 | 0.372 | 1.91535 |
Lipase activity of mix culture at different temperature using linseed oil cake as substrate
As shown in table 7, Maximum lipase activity was observed at 40℃ that was 7.69745 U/ml after 24 hr.
Table 7: Lipase activity of mix culture at different temperature using linseed oil cake as substrate
| Temp. (℃) | OD at different time interval (410nm) | |||
| 24 Hr | Activity of lipase (U/ml) | 48Hr | Activity of lipase (U/ml) | |
| 35 | 1.166 | 6.0035 | 1.189 | 6.1219 |
| 40 | 1.495 | 7.69745 | 1.313 | 6.76035 |
| 45 | 1.428 | 7.3525 | 1.244 | 6.4051 |
| 50 | 0.318 | 1.6373 | 0.262 | 1.349 |
Lipase activity of Mix culture at different temperature using sesame oil cake as substrate
As shown in table 8, Maximum lipase activity was observed at 40℃ that was 10.6731 U/ml after 24 hr.
Table 8: Lipase activity of Mix culture at different temperature using sesame oil cake as substrate
| Temp. (℃) | OD at different time interval (410nm) | |||
| 24 Hr | Activity of lipase (U/ml) | 48Hr | Activity of lipase (U/ml) | |
| 35 | 1.233 | 6.34845 | 1.196 | 6.15795 |
| 40 | 2.073 | 10.6731 | 1.881 | 9.6849 |
| 45 | 1.38 | 7.10535 | 1.244 | 6.4051 |
| 50 | 0.441 | 2.2706 | 0.416 | 2.1419 |
Lipase activity of Mix culture at different temperature using soybean oil cake as substrate
As shown in table 9, Maximum lipase activity was observed at 40℃ that was 8.2741 U/ml after 24 hr.
| Temp. (℃) | OD at different time interval (410nm) | |||
| 24 Hr | Activity of lipase (U/ml) | 48Hr | Activity of lipase (U/ml) | |
| 35 | 1.169 | 6.01895 | 1.144 | 5.89025 |
| 40 | 1.607 | 8.2741 | 1.586 | 8.166 |
| 45 | 1.549 | 7.9755 | 1.323 | 6.81185 |
| 50 | 0.149 | 0.76715 | 0.291 | 1.4983 |
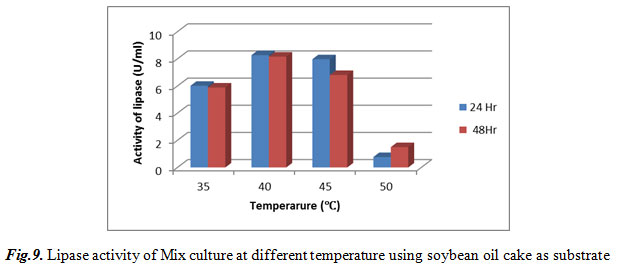 |
Figure 9: Lipase activity of Mix culture at different temperature using soybean oil cake as substrate |
Inoculum concentration
Lipase activity of mix culture at different inoculum concentration using mustard oil cake as substrate
As shown in the table 10, Maximum lipase activity is observed at 10% that was 10.19945836 U/ml after 24 hr.
Table 10: Lipase activity of mix culture at different inoculum concentration using mustard oil cake as substrate
| Inoculum concentration | OD at 24 hr (410) | |
| 24hr | Activity of lipase (U/ml) | |
| 5% | 1.743 | 8.97408174 |
| 10% | 1.981 | 10.19945836 |
| 15% | 1.856 | 9.555878204 |
Lipase activity of mix culture at different inoculum concentration using linseed oil cake as substrate
As shown in the table 11, Maximum lipase activity was observed at 10% that was 9.947174941 U/ml after 24 hr.
Table 11: Lipase activity of mix culture at different inoculum concentration using linseed oil cake as substrate
| Inoculum concentration | OD at 24 hr (410) | |
| 24hr | Activity of lipase (U/ml) | |
| 5% | 1.721 | 8.860811632 |
| 10% | 1.932 | 9.947174941 |
| 15% | 1.831 | 9.427162172 |
Lipase activity of mix culture at different inoculum concentration using sesame oil cake as substrate
As shown in the table 12, Maximum lipase activity was observed at 10% that was 9.62281054 U/ml after 24 hr.
Table 12: Lipase activity of mix culture at different inoculum concentration using sesame oil cake as substrate
| Inoculum concentration | OD at 24 hr (410) | |
| 24hr | Activity of lipase (U/ml) | |
| 5% | 1.634 | 8.412879841 |
| 10% | 1.869 | 9.62281054 |
| 15% | 1.703 | 8.768136089 |
Lipase activity of Mix culture at different inoculum concentration using soybean oil cake as substrate
As shown in table 13, Maximum lipase activity is observed at 10% 7.686921421 U/ml after 24 hr.
Table 13: Lipase activity of Mix culture at different inoculum concentration using soybean oil cake as substrate
| Inoculum concentration | OD at 24 hr (410) | |
| 24hr | Activity of lipase (U/ml) | |
| 5% | 1.321 | 6.801355122 |
| 10% | 1.493 | 7.686921421 |
| 15% | 1.398 | 7.1978005 |
Conclusion
Both strains Bacillus licheniformis MTCC 3244 and Bacillus coagulans MTCC 10305 showed the lipase activity. In mix culture experiment in case of each substrate, maximum lipase activity was obtained using mustard oil cake (10.199458 U/ml) at pH 8 maintaining at temperature 40℃ with 10% inoculum concentration after 24 hr, sesame oil cake (10.6731 U/ml) at pH 8 maintaining at temperature 40℃ with 0.5% inoculum concentration after 24 hr, linseed oil cake (9.947174941 U/ml) at pH 8 maintaining at temperature 40℃ with 10% inoculum concentration after 24 hr, and soybean oil cake (9.5716 U/ml) at pH 8 maintaining at temperature 30℃ with 0.5% inoculum concentration after 24 hr. Overall mix culture experiment, sesame oil cake gave highest lipase activity.
References
Alkan, H., Baysal, Z., Uyar, F., & Dogru, M. (2007). Production of lipase by a newly isolated Bacillus coagulans under solid-state. Applied Biochemistry and Biotechnology, 136(1), 183–192.
Asenjo, J. A., & Andrews, B. A. (2008). Mini-review Challenges and trends in bioseparations. Chemical Engineering, 120(September 2007), 117–120. https://doi.org/10.1002/jctb
Baltaci, M. O., Orak, T., Taskin, M., Adiguzel, A., & Ozkan, H. (2018). Enhancement of Amylase and Lipase Production from Bacillus licheniformis 016 Using Waste Chicken Feathers as Peptone Source. Waste and Biomass Valorization, 0(0), 0. https://doi.org/10.1007/s12649-018-0468-6
Bornscheuer, U. T. (2013). Enzymes in lipid modification: From classical biocatalysis with commercial enzymes to advanced protein engineering tools. OCL – Oleagineux Corps Gras Lipides, 20(1), 45–49. https://doi.org/10.1684/ocl.2012.0487
Chai, S. Y., Abbasiliasi, S., Lee, C. K., Ibrahim, T. A. T., Kadkhodaei, S., Mohamed, M. S., … Tan, J. S. (2018). Extraction of fresh banana waste juice as non-cellulosic and non-food renewable feedstock for direct lipase production. Renewable Energy, 126, 431–436. https://doi.org/10.1016/j.renene.2018.03.050
D’Annibale, A., Sermanni, G. G., Federici, F., & Petruccioli, M. (2006). Olive-mill wastewaters: a promising substrate for microbial lipase production. Bioresource Technology, 97(15), 1828–1833. https://doi.org/10.1016/j.biortech.2005.09.001
da S. Pereira, A., Fontes-Sant’Ana, G. C., & Amaral, P. F. F. (2019). Mango agro-industrial wastes for lipase production from Yarrowia lipolytica and the potential of the fermented solid as a biocatalyst. Food and Bioproducts Processing, 115, 68–77. https://doi.org/10.1016/j.fbp.2019.02.002
Domínguez, A., Deive, F. J., Sanromán, M. A., & Longo, M. A. (2003). Effect of lipids and surfactants on extracellular lipase production by Yarrowia lipolytica. Journal of Chemical Technology and Biotechnology, 78(11), 1166–1170. https://doi.org/10.1002/jctb.922
Falony, G., Armas, J. C., Mendoza, J. C. D., & Hernández, J. L. M. (2006). Production of extracellular lipase from Aspergillus niger by solid-state fermentation. Food Technology and Biotechnology, 44(2), 235–240. https://doi.org/10.1002/9780470015902.a0003014.pub2
Fickers, P., Marty, A., & Nicaud, J. M. (2011). The lipases from Yarrowia lipolytica: Genetics, production, regulation, biochemical characterization and biotechnological applications. Biotechnology Advances, 29(6), 632–644. https://doi.org/https://doi.org/10.1016/j.biotechadv.2011.04.005
Geoffry, K., & Achur, R. N. (2018). Optimization of novel halophilic lipase production by Fusarium solani strain NFCCL 4084 using palm oil mill effluent. Journal of Genetic Engineering and Biotechnology, 16(2), 327–334. https://doi.org/10.1016/j.jgeb.2018.04.003
Godoy, M. G., Gutarra, M. L. E., Maciel, F. M., Felix, S. P., Bevilaqua, J. V., Machado, O. L. T., & Freire, D. M. G. (2009). Use of a low-cost methodology for biodetoxification of castor bean waste and lipase production. Enzyme and Microbial Technology, 44(5), 317–322. https://doi.org/10.1016/j.enzmictec.2009.01.002
Gombert, A. K., Pinto, A. L., Castilho, L. R., & Freire, D. M. G. (1999). Lipase production by Penicillium restrictum in solid-state fermentation using babassu oil cake as substrate. Process Biochemistry, 35(1–2), 85–90. https://doi.org/10.1016/S0032-9592(99)00036-9
Gonçalves, C., Oliveira, F., Pereira, C., & Belo, I. (2012). Fed-batch fermentation of olive mill wastewaters for lipase production. Journal of Chemical Technology and Biotechnology, 87(8), 1215–1218. https://doi.org/10.1002/jctb.3738
Gupta, R., Gupta, N., & Rathi, P. (2004). Bacterial lipases: An overview of production, purification and biochemical properties. Applied Microbiology and Biotechnology, 64(6), 763–781. https://doi.org/10.1007/s00253-004-1568-8
Gutarra, M. L. E., De Godoy, M. G., Silva, J. D. N., Guedes, I. A., Lins, U., Castilho, L. D. R., & Freire, D. M. G. (2009). Lipase production and Penicillium simplicissimum morphology in solid-state and submerged fermentations. Biotechnology Journal, 4(10), 1450–1459. https://doi.org/10.1002/biot.200800298
Hasan, F., Shah, A. A., & Hameed, A. (2006). Industrial applications of microbial lipases. Enzyme and Microbial Technology, 39(2), 235–251. https://doi.org/10.1016/j.enzmictec.2005.10.016
Hasan, F., Shah, A. A., & Hameed, A. (2009). Methods for detection and characterization of lipases: A comprehensive review. Biotechnology Advances, 27(6), 782–798. https://doi.org/https://doi.org/10.1016/j.biotechadv.2009.06.001
Hasan, N. A., Nawahwi, M. Z., Yahya, N., & Othman, N. A. (2018). Special Issue.
Ismail, A. R., El-Henawy, S. B., Younis, S. A., Betiha, M. A., El-Gendy, N. S., Azab, M. S., & Sedky, N. M. (2018). Statistical enhancement of lipase extracellular production by Bacillus stratosphericus PSP8 in a batch submerged fermentation process. Journal of Applied Microbiology, 125(4), 1076–1093. https://doi.org/10.1111/jam.14023
Jaeger, K.-E., & Eggert, T. (2002). Lipases for biotechnology. Current Opinion in Biotechnology, 13(4), 390–397. https://doi.org/https://doi.org/10.1016/S0958-1669(02)00341-5
Jain, R., & Naik, S. N. (2018). Adding value to the oil cake as a waste from oil processing industry: Production of lipase in solid state fermentation. Biocatalysis and Agricultural Biotechnology, 15, 181–184. https://doi.org/10.1016/j.bcab.2018.06.010
Kapoor, M., & Gupta, M. N. (2012). Lipase promiscuity and its biochemical applications. Process Biochemistry, 47(4), 555–569. https://doi.org/https://doi.org/10.1016/j.procbio.2012.01.011
Long, Z. De, Xu, J. H., & Pan, J. (2007). Significant improvement of Serratia marcescens lipase fermentation, by optimizing medium, induction, and oxygen supply. Applied Biochemistry and Biotechnology, 142(2), 148–157. https://doi.org/10.1007/s12010-007-0023-6
Martínez-Herrera, J., Siddhuraju, P., Francis, G., Dávila-Ortíz, G., & Becker, K. (2006). Chemical composition, toxic/antimetabolic constituents, and effects of different treatments on their levels, in four provenances of Jatropha curcas L. from Mexico. Food Chemistry, 96(1), 80–89. https://doi.org/https://doi.org/10.1016/j.foodchem.2005.01.059
Mendes, A. A., Oliveira, P. C., & de Castro, H. F. (2012). Properties and biotechnological applications of porcine pancreatic lipase. Journal of Molecular Catalysis B: Enzymatic, 78, 119–134. https://doi.org/https://doi.org/10.1016/j.molcatb.2012.03.004
Mitchell, D. A., Berovic, M., & Krieger, N. (2000). Biochemical Engineering aspects solid state bioprocessing. Bioprocessing. 68, 3–4.
Rigo, E., Ninow, J. L., Di Luccio, M., Vladimir Oliveira, J., Polloni, A. E., Remonatto, D., … Treichel, H. (2010). Lipase production by solid fermentation of soybean meal with different supplements. LWT – Food Science and Technology, 43(7), 1132–1137. https://doi.org/10.1016/j.lwt.2010.03.002
Rocha, M. A., Padez, C., & Morais, M. H. X. de. (1998). Urbanização e idade da menarca na população portuguesa: evolução secular (1880- 90 a 1980). Antropologia Portuguesa, 15(5), 59–75. https://doi.org/10.5897/SRE11.2023
Salihu, A., Alam, M. Z., AbdulKarim, M. I., & Salleh, H. M. (2012). Lipase production: An insight in the utilization of renewable agricultural residues. Resources, Conservation and Recycling, 58, 36–44. https://doi.org/10.1016/j.resconrec.2011.10.007
Salihu, A., Bala, M., & Alam, M. Z. (2016). Lipase production by Aspergillus niger using sheanut cake: An optimization study. Journal of Taibah University for Science, 10(6), 850–859. https://doi.org/10.1016/j.jtusci.2015.02.011
Saravanan, A. N., Suchitra, N., & Dhandayuthapani, K. (2007). Role of Saturated Fatty Acids in Lipase Production –. Journal of Food Biochemistry, 31(2007), 748–756.
Selvakumar, P., & Sivashanmugam, P. (2017). Optimization of lipase production from organic solid waste by anaerobic digestion and its application in biodiesel production. Fuel Processing Technology, 165, 1–8. https://doi.org/10.1016/j.fuproc.2017.04.020
Sharma, R., Chisti, Y., & Chand, U. (2011). Live – Wednesday football. Bbc, 19, 627–662. https://doi.org/10.1016/S0734-9750(01)00086-6
Singhania, R. R., Soccol, C. R., & Pandey, A. (2008). Application of Tropical Agro-industrial Residues as Substrate for Solid-state Fermentation Processes. In A. Pandey, C. R. Soccol, & C. Larroche (Eds.), Current Developments in Solid-state Fermentation (pp. 412–442). https://doi.org/10.1007/978-0-387-75213-6_18
Sreelatha, B., Koteswara Rao, V., Ranjith Kumar, R., Girisham, S., & Reddy, S. M. (2017). Culture conditions for the production of thermostable lipase by Thermomyces lanuginosus. Beni-Suef University Journal of Basic and Applied Sciences, 6(1), 87–95. https://doi.org/10.1016/j.bjbas.2016.11.010
Suci, M., Arbianti, R., & Hermansyah, H. (2018). IOP Conference Series: Earth and Environmental Science Lipase production from Bacillus subtilis with submerged fermentation using waste cooking oil Lipase production from Bacillus subtilis with submerged fermentation using waste cooking oil. 105, 12126. https://doi.org/10.1088/1755-1315/105/1/012126
Sun, J., Yu, B., Curran, P., & Liu, S.-Q. (2013). Lipase-catalysed ester synthesis in solvent-free oil system: Is it esterification or transesterification? Food Chemistry, 141(3), 2828–2832. https://doi.org/https://doi.org/10.1016/j.foodchem.2013.05.109
Sun, S. Y., & Xu, Y. (2008). Solid-state fermentation for ‘whole-cell synthetic lipase’ production from Rhizopus chinensis and identification of the functional enzyme. Process Biochemistry, 43(2), 219–224. https://doi.org/https://doi.org/10.1016/j.procbio.2007.11.010
Taskin, M., Ucar, M. H., Unver, Y., Kara, A. A., Ozdemir, M., & Ortucu, S. (2016). Lipase production with free and immobilized cells of cold-adapted yeast Rhodotorula glutinis HL25. Biocatalysis and Agricultural Biotechnology, 8, 97–103. https://doi.org/10.1007/978-3-319-63754-9_4
Turati, D. F. M., Almeida, A. F., Terrone, C. C., Nascimento, J. M. F., Terrasan, C. R. F., Fernandez-Lorente, G., … Carmona, E. C. (2019). Thermotolerant lipase from Penicillium sp. section Gracilenta CBMAI 1583: Effect of carbon sources on enzyme production, biochemical properties of crude and purified enzyme and substrate specificity. Biocatalysis and Agricultural Biotechnology, 17, 15–24. https://doi.org/10.1016/j.bcab.2018.10.002
Vasiee, A., Behbahani, B. A., Yazdi, F. T., & Moradi, S. (2016). Optimization of the production conditions of the lipase produced by Bacillus cereus from rice flour through Plackett-Burman Design (PBD) and response surface methodology (RSM). Microbial Pathogenesis, 101, 36–43. https://doi.org/10.1016/j.micpath.2016.10.020
Yu, M., Wen, S., & Tan, T. (2010). Enhancing production of Yarrowia lipolytica lipase Lip2 in Pichia pastoris. Engineering in Life Sciences, 10(5), 458–464. https://doi.org/10.1002/elsc.200900102
Zouaoui, B., & Bouziane, A. (2012). Isolation, Optimisation and Purification of Lipase Production by Pseudomonas Aeruginosa. Journal of Biotechnology & Biomaterials, 01(07), 10–13. https://doi.org/10.4172/2155-952x.1000120


