1Department of Biotechnology, Ambala College of Engineering and Applied Research, Devsthali-133101, Ambala, Haryana, India
2Department of Biotechnology, Maharishi Markandeshwar University Sadopur Ambala -134007, Haryana, India
3Department of Botany and Biotechnology, Dayanand College, Hisar, 125001, Haryana, India
4Department of Biotechnology, Chaudhary Devi Lal University, Sirsa-125055, Haryana, India
Corresponding author Email: mukesh.biotech@gmail.com
Article Publishing History
Received: 22/12/2018
Accepted After Revision: 21/03/2019
In the present study, a tannin acyl hydrolase producer bacterium Klebsiella pneumoniae (GenBank Accession Number KP715242) isolated from the soil was used for enhanced tannase production. An attempt was carried out to develop an efficient tannase producing strain by subjecting this parent strain (K. pneumoniae KP715242) to various mutagenic agents viz. heat, UV irradiations, NTG (N-methyl-N-nitro- N-nitrosoguinidine) and MMS (methyl methane sulfonate). Two most effi ciently growing colonies per treatment were selected at 5–10% survival rate for each treatment. These mutants were assessed for enzyme production under submerged fermentation at preoptimized culture conditions i.e. 5.2 pH, 34.97°C temperature, 103.3 4 rpm agitation speed and 91.34 h of incubation time. Among these, the maximum tannase production (0.075 U/ml) was observed for the mutant U2. The most efficient mutant strain U2 had nearly 1.15 fold more enzyme activity as compared to the wild type strain (0.065 U/ml) after 90 h of incubation time under similar culture conditions. The maximum enzyme production in this mutant strain was 0.063 U/mL after an incubation time of 75 h as compared to wild type strain that had an incubation period of 90 h for a maximum enzyme activity of 0.065 U/mL. The results indicated that the mutated strain U2 exhibited higher tannase activity as well as less incubation as compared to the wild type strain.
Klebsiella Pneumoniae, Methyl Methane Sulfonate, Mutagenesis, N-Methyl-N-Nitro-N-Nitrosoguinidine, Strain Improvement, Tannase
Kumar M, Rajesh, Srivastava V, Salar R. K. Upgradation of Tannase Production by Klebsiella Pneumoniae KP715242 Through Heat, UV, NTG and MMS Induced Mutagenesis for Enhanced Tannase Activity. Biosc.Biotech.Res.Comm. 2019;12 (1).
Kumar M, Rajesh, Srivastava V, Salar R. K. Upgradation of Tannase Production by Klebsiella Pneumoniae KP715242 Through Heat, UV, NTG and MMS Induced Mutagenesis for Enhanced Tannase Activity.. Biosc.Biotech.Res.Comm. 2019;12(1). Available from: https://bit.ly/2wtLpn7
Copyright © Kumar et al., This is an open access article distributed under the terms of the Creative Commons Attribution License (CC-BY) https://creativecommns.org/licenses/by/4.0/, which permits unrestricted use distribution and reproduction in any medium, provide the original author and source are credited.
Introduction
Tannins are water soluble polyphenolic compounds of varying molecular weights. These are extensively dispersed in the plant kingdom and are the fourth most abundant plant component after cellulose, hemicellulose and lignin. Tannins own several imperative biological properties, like defense against fungal, bacterial, and viral diseases. Tannins defend the susceptible parts of the plants from microbial attack by deactivating the attacking microbial extracellular enzymes (Aguilar and Sanchez, 2001). Tannins interact promptly with proteins and other molecules to make them indigestible, and adversely affect the nutritive value of animal food (Bhat et al., 1993). High concentration of tannic acid in soil has several detrimental effects like plant vegetation, cropping systems and low production yield. Moreover, tannin also causes various nutritional and processing problems like protein indigestibility, inhibition of enzymatic reactions and essential microbial processes such as those necessary for beer brewing. These undesirable properties of tannin are responsible for reduced voluntary feed intake, ineffi cient digestion and low animal productivity (Jana et al., 2014).
Tannin acyl hydrolase (tannase) causes the degradation of hydrolysable tannins because of its hydrolyzing ability for the ester and the depside bond with subsequent release of glucose, gallic acid and several galloyl esters of glucose (Haslam and Stangroom, 1996; Govindarajan et al., 2019). Tannase belongs to the esterase superfamily and has been shown to be intracellular as well as extracellular (Aguilar et al., 2007; Banerjee and Mahapatra, 2012). This enzyme has found numerous potential in different industrial aspects like food, brewing, beverages and, pharmaceuticals. It has been profoundly used for the preparation of instant tea, clarifi – cation of various fruit juices with other beverages and more importantly in the gallic acid production (Belmares et al., 2004; Mohapatra et al., 2009; Madeira et al., 2011; Chavez-Gonzalez et al., 2012, Ghosh and Mandal, 2015; Fathy et al., 2018; Aharwar et al., 2018).
Tannase has been reported to be present in plants, animals and microorganisms, with maximum production from the latter. It is synthesized by various microbes like bacteria, fungi, and yeast. However, maximum research work has been done in case of fungi. Nevertheless, fungal applications at commercial level are largely discouraged owing to its rather low growth rate coupled with relatively complex genome. On the other hand, growth potential of bacteria is very high and are comparatively easier to be handled at the genomic level. Apart from this, the bacteria also owe the ability to endure extreme temperature conditions, thereby exhibiting greater thermo-stability (Kumar et al., 2015). Moreover, bacterial tannase can also degrade and hydrolyze natural tannins and tannic acid very effi ciently (Deschamps et al., 1983).
Production of tannase from microorganisms can be achieved through different methods like liquid surface, submerged (Smf), solid state fermentation (SSF) and modifi ed solid-state fermentation (Belmares et al., 2004). Submerged fermentation is usually considered as more valuable because of easiness of sterilization, and process control to engineer in these systems (Mahapatra et al., 2005; Jana, 2013; Govindarajan et al., 2019). In view of the immense potential of tannase, large number of efforts have been carried out to make the production and purification of this enzyme more efficient. These efforts include the exploration of novel sources of tannase (Mahapatra and Banerjee, 2009; Chhokar et al., 2010; Zakipour et al., 2013) search of new and more effective fermentation systems (Purohit et al., 2006), statistical optimization of different culture conditions (Raaman et al., 2010; Aharwar et al., 2018), development of more effi cient procedures for purification and recovery of tannase and enhanced tannase production through mutagenesis and recombinant microorganism (Curiel et al., 2009; Liu et al., 2018).
The present study represents a part of these efforts with an aim to produce a more efficient tannase producing mutant bacterial strain. In this study, we have made an attempt to enhance tannase production by different mutagenic approaches from culture of Klebsiella pneumoniae KP715242
Materials and Methods
Isolation and Identification of Tannase Producing Bacterial Strain
A number of soil samples collected from different regions of Ambala, India were screened for the isolation of tannase positive bacterial strains. Each soil sample (1.0 gm) was suspended in 50 ml minimal medium containing K2HPO4:0.5g/l, KH2PO4:0.5g/l, MgSO4:2.0g/l, CaCl2: 1.0g/l and NH4Cl: 3.0g/l supplemented with 1% tannic acid (pH 5.5) and incubated at 37°C on rotary shaker at 150 rpm for 24 hours (Osawa, 1990). A portion of each culture was transferred to fresh medium and cultured again under same conditions until pure cultures were obtained. The strains were maintained on nutrient agar slants in refrigerator at 4°C by regular transfer. All the pure isolates were assessed for tannic acid degradation using through visual reading method and zone of hydrolysis (Kumar et al., 2015a).
Bacterial strain with maximum tannase activity (TAH 10) was identified on the basis of its morphological, physiological, biochemical characteristics and 16S rRNA gene sequence.
Tannase Assay
For the tannase assay, 200 μl enzyme solution was mixed with 300μl of 1.0% (w/v) tannic acid prepared in 0.2 M acetate buffer of pH 5.5. The mixture was incubated for 40 minutes at 40°C, thereafter the reaction was stopped by the addition of 3.0 ml bovine serum albumin (1 mg/ mL) that caused the precipitation of the residual tannic acid. The heat denatured enzyme was used as a control and was processed in similar way. The reaction mixture containing tubes were centrifuged at 7,000 x g for 10 minutes followed by the dissolution of the precipitates in 3.0 ml of SDS–triethanolamine (1% w/v) solution. Thereafter, 1.0 ml of FeCl3 reagent (0.13 M) was added and the mixture was incubated for 15 minutes to get the color stabilized. The enzyme activity was measured by reading the absorbance of both the sample and control tubes at 530 nm against the blank (without tannic acid). One unit of the tannase was defi ned as the amount of enzyme, which is able to hydrolyze 1 mM of substrate tannic acid in 1 minute under assay conditions (Mondal et al., 2001; Kumar et al., 2016).
Formulation of Cell Suspension
For the preparation of the cell suspension, a small portion of 24 hours grown culture of K. pneumoniae was added in 50 mL nutrient broth followed by subsequent incubation at 37 °C for 12 hours under shaking conditions (110 rpm). A cell biomass pellet was obtained by centrifuging 10 ml inoculated broth at 9000 x g for 10 min at 4°C. The cell suspension (approximately 107-8 cells/ml) was prepared by dissolving this pellet in 10 ml sterilized saline and was used for further studies.
Heat Induced Mutagenesis
In order to carry out heat induced mutagenesis, 1.0 ml of cell suspension was transferred to a sterilized test tube and was heated at 60 °C for different time intervals ranging from 20 to 70 minutes. The cell suspension was serially diluted to obtain a final dilution of 10-6/ml. After the dilution, 100 μl of heat treated cell suspension was laid over the nutrient agar plates supplemented with 2% tannic acid under sterile conditions. All the plates (20, 30, 40, 50, 60 and 70 min) were processed in the same manner. After keeping all the plates in dark for 30 minutes, they were incubated for 48 hours at 37°C. Subsequently, the survival percentage was determined over the control (plate containing unheated cell suspension).
Uv Induced Mutagenesis
For the induction of UV induced mutagenesis, 1.0 ml of the appropriately diluted cell suspension was taken separately in different Petri plates. All of these plates were exposed to UV radiations for different periods ranging from 10 to 60 minutes by maintaining a distance of about 10 cm from the source of UV radiations. After the desired time period, the plates were given dark treatment for 30 minutes, plated on nutrient agar plates provided with 2% tannic acid and incubated for 48 hours at 37°C. The survival percentage of bacterial cells was calculated over the untreated control plate.
Ntg (N-Methyl N-Nitro N-Nitrosoguanidine) and Mms (Methyl Methane Sulfonate) Induced Mutagenesis
To observe the effect of NTG and MMS mutagens, the bacterial cell suspension and NTG (1 mg/ml) and MMS (100 μl/ml) solutions were mixed separately in a ratio of 1:1 in corresponding test tubes excluding the control tube. This mixture was incubated at 37 ºC for different time periods ranging from 10-60 min. After the desired period of treatment, mixture solution containing treated bacterial cells was centrifuged at 8000 x g for 10 min at 4 ºC. The pellet obtained was washed twice with sterilized normal saline and plated on nutrient agar plates supplemented with 2% tannic acid. The plates were given dark treatment for 2 hours and then incubated at 37 ºC for 48 hours. The cell survival percentage was determined using the plate containing the untreated cell suspension as control.
Comparison of Tannase Production Between Mutant Strains From Each Treatment
For the isolation and identification of hyper tannase producer strain, two colonies from around 5% survival rate were selected for each treatment. The selected isolates were used for the production of extracellular tannase in submerged conditions. Tannase production was carried out in 250 ml Erlenmeyer’s fl ask with 50 ml of fermentation medium(0.5 g/l K2HPO4, 0.5 g/l KH2PO4, 2.0 g/l MgSO4, 1.0 g/l CaCl2 and 3.0 g/l NH4Cl) enriched with 1% tannic acid under optimized conditions. After regular intervals of 15, 30, 45, 60, 75, 90 and 105 hours, 1.0 ml of the fermented medium was aseptically removed and centrifuged at 10,000 rpm for 20 min at 4°C. The obtained supernatant (crude enzyme) was used for the estimation of tannase activity.
Results and Discussion
Isolation and Screening of Bacteria
A total of 13 pure isolates out of 20 were able to show significant growth on nutrient agar plates supplemented with 1% tannic acid. Out of these 13 isolates, only 6 isolates were found to be positive for visual detection method. However, only TAH 10 was able to exhibit distinct zone of hydrolysis on the agar plates (Fig. 1) with maximum tannase activity of 0.02 U/ml amongst all 6 tested strains (Table 1). Tannase production from this wild type strain was maximized by optimizing the various culture conditions. Under these optimum conditions (5.2 pH, 34.97 °C temperature, 103.34 rpm agitation speed and 91.34 hours of incubation time) this strain exhibited 0.065 U/ml of tannase in comparison to 0.025 U/ml obtained under un-optimized conditions (Kumar et al., 2015a).
 |
Figure 1: Nutrient agar plate showing tannase-producing bacterium TAH 10 after 4 days incubation |
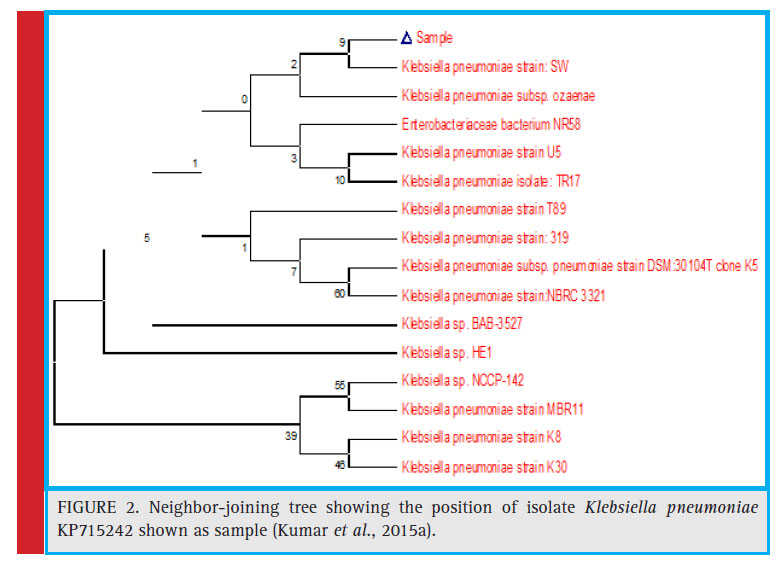 |
Figure 2: Neighbor-joining tree showing the position of isolate Klebsiella pneumoniae KP715242 shown as sample (Kumar et al., 2015a). |
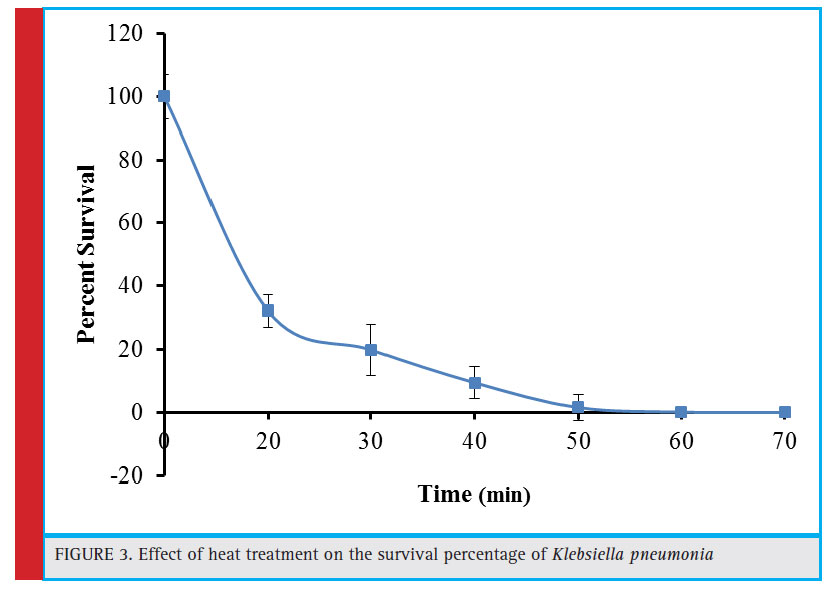 |
Figure 3: Effect of heat treatment on the survival percentage of Klebsiella pneumonia |
Identification of Bacterial Isolate
On the basis of morphological characterization, the bacterial strain TAH 10 was found to be a gram negative, non-motile, rod shaped bacterium. Amplifi cation of 16S rDNA and its subsequent gene sequence was used to construct the phylogenetic tree (Fig. 2). The 16S rDNA sequence of the strain TAH 10 has been deposited in the NCBI GeneBank (Accession Number KP715242).
Development of Efficient Tannase Producing Bacterial Strain Through Mutagenesis
For the development of hyper tannase producing bacterial strain, the parental isolate Klebsiella pneumoniae was subjected to various mutagenic treatments like heat treatment, ultra-violet (UV) radiation, N-methyl N-nitro N-nitrosoguanidine (NTG) treatment and methyl methane sulfonate (MMS) treatment. The action of such kind of mutagenic treatments is random in nature that may also lead to mutations in the desired genes resulting in the impairment of essential functions culminating in the death of the cells.
Influence of Heat on the Survivability of Klebsiella Pneumoniae
The heat treatment (60°C) associated survival percentage of the bacterial cells is depicted in figure 3. The survival percentage declined to 32.20% after initial 20 minutes of heat treatment. There was a progressive decrease in the survival rate with the passage of time and the culture was almost lost after 50 minutes of treatment. The mechanism of heat action involves generation of an oxidative stress in the cells and such heat generated stress has been reported to be lethal at 50°C in Saccharomyces cerevisiae (Davidson and Schiestl, 2000). The other heat associated deleterious effects include mutations, recombination, and mitochondrial membrane disruption which are thought to cause decreased sulfhydryl content. Similarly, the co-culture of Aspergillus foetidus and Rhizopus oryzae was completely eliminated after 60 minute exposure of heat treatment (Purohit et al., 2006). Zakipour et al. (2013) reported enhanced production of tannase from Penicillium sp. EZ-ZH190 through induced mutation. It was observed that after 10 minute exposure of heat treatment at 60°C, 8.5×104 colonies (17% of the initial number) were obtained that declined gradually with the increase in exposure time and became nil after exposure of 40, 50, and 60 minutes.
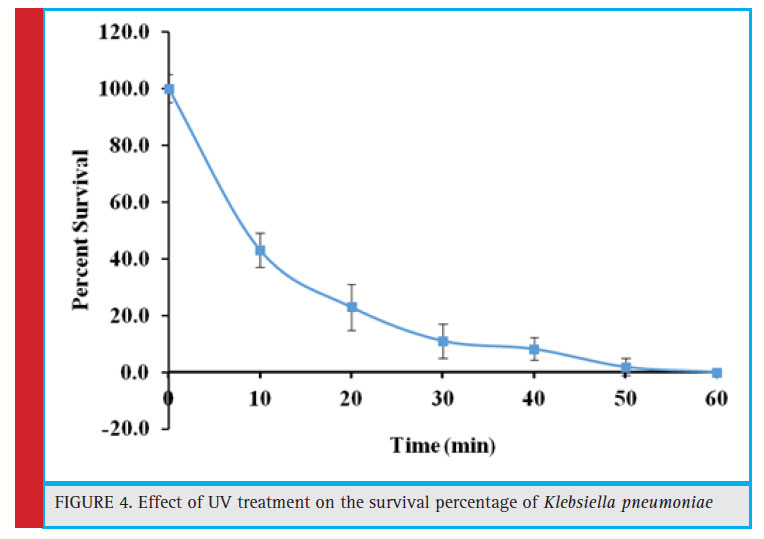 |
Figure 4: Effect of UV treatment on the survival percentage of Klebsiella pneumoniae |
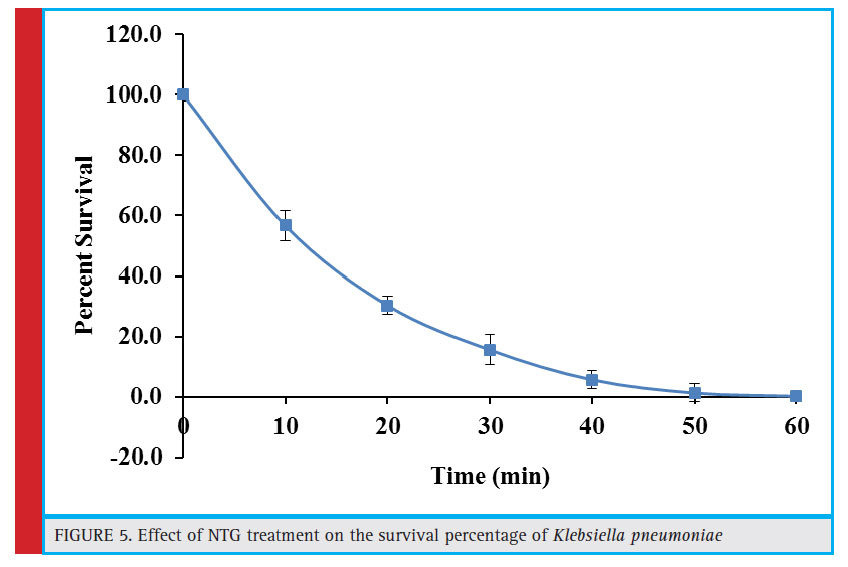 |
Figure 5: Effect of NTG treatment on the survival percentage of Klebsiella pneumoniae |
Influence of Ntg (N-Methyl N-Nitro N-Nitrosoguanidine) Mutagens on the Survivability of Klebsiella Pneumoniae
NTG mutagen induces repetitive mutations and strongly increase their frequency. It precisely interacts with DNA and is considered as UV-mimetic agent. In addition, NTG may also form nitrosoguanidine group containing reaction products upon reaction with organic ions. It may inhibit the synthesis of functional proteins or could affect protein synthesis by causing mistakes during translation (Vorobjeva, 2013). These inhibitory effects of NTG have also been observed in the present study. After initial exposure of 10 minutes, the percent survival rate of Klebsiella pneumoniae was reduced to 56.7%. The survival rate declined steadily with increase in the exposure time and the culture was nearly eliminated after 50 minutes of treatment (Fig. 5). A similar trend has been observed in case of co-culture of Aspergillus foetidus and Rhizopus oryzae where 60 minute exposure to NTG completely eliminated the culture (Purohit et al., 2006).
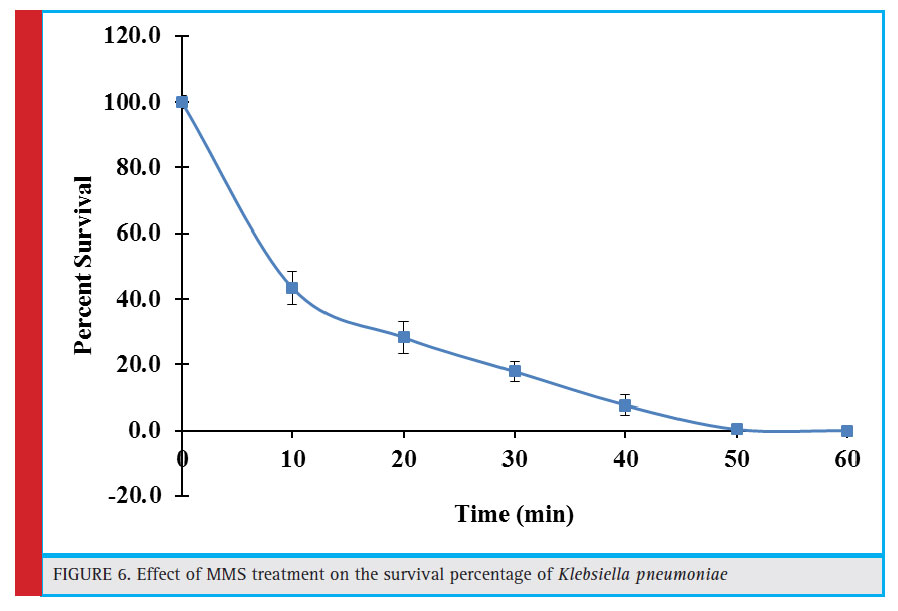 |
Figure 6: Effect of MMS treatment on the survival percentage of Klebsiella pneumoniae |
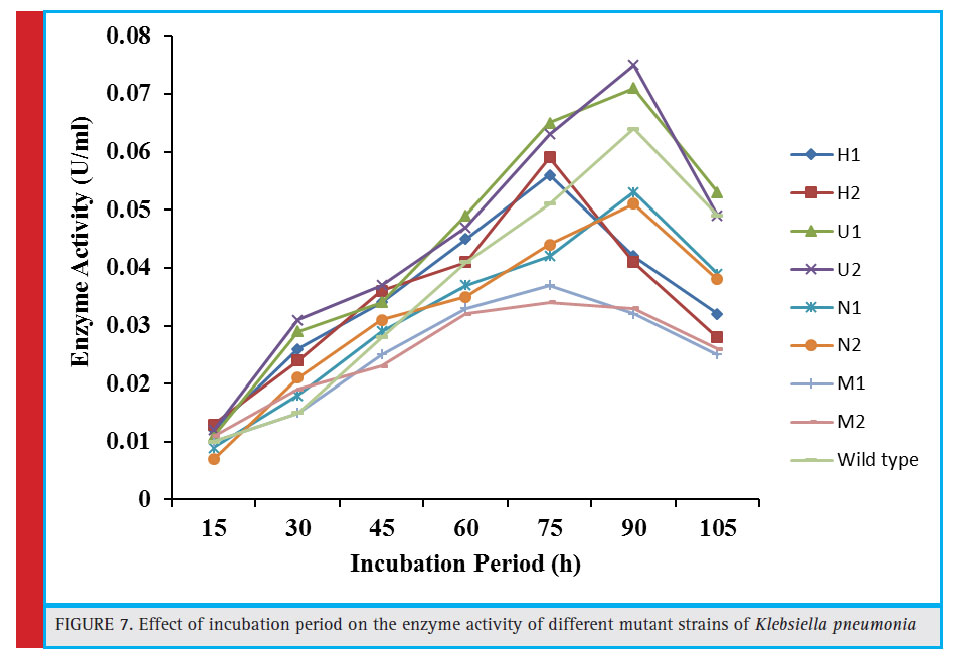 |
Figure 7: Effect of incubation period on the enzyme activity of different mutant strains of Klebsiella pneumonia |
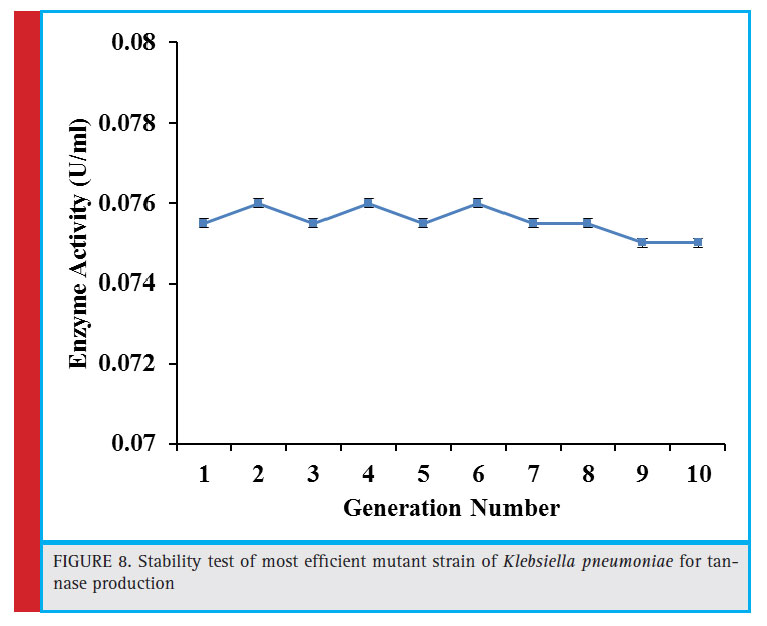 |
Figure 8: Stability test of most effi cient mutant strain of Klebsiella pneumoniae for tannase production |
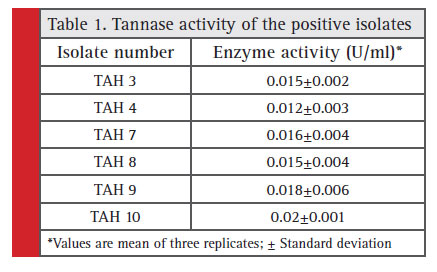 |
Table 1: Tannase activity of the positive isolates |
Effect of Mms (Methyl Methane Sulfonate) Treatment on the Survivability of Klebsiella Pneumoniae
Methylmethane sulphonate (MMS), is a type of alkylating agent and possess cytotoxic and mutagenic properties. It primarily methylates purine bases leading to various adduct formation in the DNA. Most of the methylated bases are toxic to the cell, hamper DNA replication and can also be a source of base pair substitutions mutations. In this report, initial 10 minute exposure to MMS resulted in 43.4% survival rate which declined gradually as the exposure time increased and was practically lost as the exposure time reached 50 minutes (Fig. 6).
Selection of Most Efficient Tannase Producing Mutant Strain
From each mutagenic treatments, two most efficiently growing colonies per treatment were selected from 5–10% survival rate from the nutrient agar plates. These bacterial colonies were carefully chosen from the plates having 40-minute exposure to heat (H1 and H2), UV (UV1 and UV2), NTG (N1 and N2) and MMS (M1 and M2) treatments.
For the isolation and selection of most efficient tannase producing mutant strains, these mutants were further assessed for enzyme production under submerged fermentation at preoptimized culture conditions i.e. 5.2 pH, 34.97°C temperature, 103.34 rpm agitation speed and 91.34 hour of incubation time as reported in our previous study (Kumar et al., 2015a). Among these, the maximum tannase production (0.075 U/ml) was witnessed in mutant U2 followed by U1, H2, and H1, producing 0.065, 0.059 and 0.056 U/ml, respectively (Fig. 7). It was found that the U2 was the most effi cient tannase producing mutant, giving approximately 1.15 fold more enzyme over the wild type strain (0.065 U/ml) after 90 hours of incubation time under similar culture conditions. The maximum tannase in this mutant strain was 0.065 U/mL with an incubation period of 75 hours as compared to
wild strain where the incubation period was 90 hours in order to exhibit the same enzyme activity. Moreover, for the mutant strain U2 generated by 40 minute UV treatment, the incubation time lowered from decreased 90 hours to 75 hours to exhibit the same enzyme activity (0.065 U/mL ) as obtained in case of wild culture.
The present results are in accordance to the previous report of enhanced tannase production through mutagenesis highlighting 1.21 fold increase in tannase production from mutant SCPR 337, produced by 60-minute heat treatment (Purohit et al., 2006). In addition, the incubation period also decreased to 30 hours from 48 hours for maximum enzyme production as compared to the wild co-culture of Aspergillus foetidus and Rhizopus oryzae. Similarly, Zakipour et al. (2013) also observed that the incubation time of the mutant B named Penicillium sp. EZ-ZH290 (generated by 30 minute heat treatment at 60 °C) for maximum tannase production, decreased from 96 hours to 84 hours retaining the same tannase activity of 4.3 U/ml in comparison to the wild culture.
The best performing mutant strain U2 was assessed for its tannase producing capability for 10 generations. Figure 8 shows that this mutant maintained the production efficiency endorsing the stability and heritability of the mutation.
Conclusion
The tannase producer bacterium K. pneumoniae (Gen- Bank Accession Number KP715242) used in the present study was isolated from soil and was subsequently identified at the molecular level through 16S ribosomal RNA gene sequence. Induction of mutations through various mutagenic agents and screening of commercially significant microorganisms are crucial processes for the efficacious development of various microbial strains required for fermentation industry. Enhanced tannase production through mutagenesis has been reported in case of fungi but till date, there is no report on strain improvement in case of bacteria. Therefore, mutagenesis was induced for the first time in the bacterial parent strain Klebsiella pneumoniae through various mutagenic treatments in an attempt for the development of more efficient tannase producing mutant bacterial strain. Amongst different mutagenic agents, UV-radiations were found to be most effective in generating more efficient tannase producer mutant strain. In comparison to the wild type strain, the improved mutated strain delivered enhanced tannase production coupled with reduced incubation period. Moreover, the most efficient mutant strain retained its tannase production efficacy for 10 generations reflecting the stability and inheritability of the mutation. Such desirable properties of this mutant strain could be exploited for commercial production of tannase at large scale with reduced cost.
Acknowledgements
The authors thank Chairperson, Department of Biotechnology, Chaudhary Devi Lal University, Sirsa, Haryana, India for providing necessary laboratory facilities to carry out this work.
Funding
This research did not receive any specific grant from funding agencies in the public, commercial, or not-for profit sectors.
References
Aguilar C.N., Rodríguez R., Gutiérrez-Sánchez G., Augur C., Favela-Torres E., Prado-Barragan L.A., Ramírez-Coronel A., Contreras-Esquivel J.C. (2007) Microbial tannases: advances and perspectives. Applied Microbiology and Biotechnology 6: 47–59.
Aguilar C.N., Sanchez G.G. (2001) Review: Sources properties, applications and potential uses of tannin acyl hydrolase. International Journal of Food Science & Technology 7(5): 373–382.
Aharwar A., Parihar D.K. (2018) Tannases: Production, Properties, Applications. Biocatalysis and Agricultural Biotechnology 15: 322-334.
Banerjee D., Mahapatra S. (2012) Fungal tannase: a journey from strain isolation to enzyme applications. Dynamic Biochemistry, Process Biotechnology and Molecular Biology 6:49–60.
Belmares R., Contreras-Esquivel J.C., Rodriguez-Herrera R., Coronel A.R., Aguilar C.N. (2004) Microbial production of tannase: an enzyme with potential use in food industry. Lebensmittel- Wissenschaft und -Technologie 37: 857-864.
Bhat T.K., Singh B. Sharma O.P. (1998) Microbial degradation of tannins – a current perspective. Biodegradation 9: 343– 57.
Chavez-Gonzalez M., Rodrigues-Durán L.V., Balagurusamy N., PradoBarragán A., Rodrigues R., Conheras J.C., Aguilar G.N. (2012) Biotechnological advances and challenges of tannase: an overview. Food and Bioprocess Technology 5(2): 445-459.
Chhokar V., Seema, Beniwal V., Salar R.K., Nehra K.S., Kumar A., Rana J.S. (2010) Purifi cation and characterization of extracellular
tannin acylhydrolase from Aspergillus heteromorphus MTCC 8818. Biotechnology and Bioprocess Engineering 15: 793–799.
Curiel J.A., Rodriguez H., Acebron I., Mancheno J.M., Rivas de las B., Munoz R. )2009) Production and physicochemical properties of recombinant Lactobacillus plantarum tannase. Journal of Agricultural and Food Chemistry 57: 6224–6230.
Davidson J.F., Schiestl R.H. (2000) Mis-targeting of multiple gene disruption constructs containing hisG. Current Genetics 38(4): 188-90.
Deschamps A.M., Otuk G., Lebeault J.M. (1983) Production of tannase and degradation of chestnut tannin by bacteria. Journal of Fermentation Technology, 61: 55–59.
Fathy S.A., Mahmoud A.E., Rashad M.M., Ezz M.K., Mohammed A.T. (2018) Improving the nutritive value of olive pomace by solid state fermentation of Kluyveromyces marxianus with simultaneous production of gallic acid. International Journal of Recycling of Organic Waste in Agriculture 7(2): 135-141.
Ghosh K., Mandal S. (2015) Nutritional evaluation of groundnut oil cake in formulated diets for rohu, Labeo rohita (Hamilton) fingerlings after solid state fermentation with a tannase producing yeast, Pichia kudriavzevii (GU939629) isolated from fish gut. Aquaculture Reports 2: 82–90.
Govindarajan R.K., Mathivanan K., Rameshkumar N., Shyu D.J.H., Kayalvizhi N. (2019) Purifi cation, structural characterization and biotechnological potential of tannase enzyme produced by Enterobacter cloacae strain 41 Process Biochemistry. 77: 37-47.
Haslam E., Stangroom J.E. (1996) The esterase and depsidase activities of tannase. Journal of Biochemistry 99(1): 28–31.
Jana A, Halder S.K., Banerjee A, Paul T, Pati B.R., Mondal K.C., Mohapatra P.K.D. (2014) Biosynthesis, structural architecture and biotechnological potential of bacterial tannase: A molecular advancement, Bioresource Technology 157: 327–340.
Jana A., Maity C., Halder S.K., Das A., Pati B.R., Mondal K.C., Mohapatra P.K.D. (2013) Structural characterization of thermostable, solvent tolerant, cytosafe tannase from Bacillus subtilis PAB2. Biochemical Engineering Journal 77: 161–170.
Kumar M., Amrinder S., Beniwal V., Salar R.K. (2016) Improved production of tannase by Klebsiella pneumoniae using Indian gooseberry leaves under submerged fermentation using Taguchi approach. AMB Express 6: 46.
Kumar M., Beniwal V., Salar R.K. (2015) Purifi cation and characterization of a thermophilic tannase from Klebsiella pneumoniae KP715242. Biocatalysis and Agricultural Biotechnology 4: 745–751.
Kumar M., Rana S., Beniwal V., Salar R.K. (2015a)Optimization of tannase production by a novel Klebsiella pneumoniae KP715242 using central composite design. Biotechnology Reports 7: 128–134.
Liu F., Wang B., Ye Y., Pan L. (2018) High level expression and characterization of tannase tan7 using Aspergillus niger SH-2 with low-background endogenous secretory proteins as the host. Protein Expression and Purifi cation 144: 71–75.
Madeira J.J.V., Macedo J.A., Macedo G.A. (2011) Detoxifi cation of castor bean residues and the simultaneous production of tannase and phytase by solid-state fermentation using Paecilomyces variotii. Bioresource Technology102: 7343–7348.
Mahapatra K., Nanda R.K., Bag S.S., Banerjee R., Pandey A., Szakacs G. (2005) Purifi cation, characterization and some studies on secondary structure of tannase from Aspergillus awamori nakazawaI. Process Biochemistry 40(10): 3251–3254.
Mahapatra S., Banerjee D. (2009) Extracellular tannase production by endophytic Hyalopus sp. The Journal of General and Applied Microbiology 55(3): 255-259.
Mohapatra P.K.D., Maity C., Rao R.S., Pati B.R., Mondal K.C. (2009) Tannase production by Bacillus licheniformis KBR6: Optimization of submerged culture conditions by Taguchi DOE methodology. Food Research International 42: 430–435.
Mondal K.C., Banerjee D., Jana M., Pati B.R. (2001) Colorimetric assay for determination of tannin acyl hydrolase (E.C.3.1.1.20) activity. Analytical Biochemistry 295: 168–171.
Osawa R. (1990) Formation of a clear zone on tannin-treated brain heart infusion agar by a Streptococcus sp. isolated from feces of koalas. Applied and Environmental Microbiology 56: 829-831.
Purohit J.S., Dutta J.R., Nanda R.K., Banerjee R. (2006) Strain improvement for tannase production from co-culture of Aspergillus foetidus and Rhizopus oryzae. Bioresource Technology 97: 795-801.
Raaman N., Mahendran B., Jaganathan C., Sukumar S., Chandrasekaran V. (2010) Optimisation of extracellular tannase production from Paecilomyces variotii. World Journal of Microbiology and Biotechnology 26: 1033–1039.
Vorobjeva L. (2013) Propionibacteria. Springer Science & Business Media. Zakipour M.E., Hamidi-Esfahani Z., Sahari M.A., Azizi M.H.
(2013) Improvement of strain Penicillium sp. EZZH 190 for tannase production by induced mutation. Applied Biochemistry and Biotechnology 17(6): 1376-1389.


