Department of Plant Molecular Biology and Biotechnology, Indira Gandhi Agricultural University, College of Agriculture, Raipur, C.G, 492012, India
Corresponding author Email: ghchandel@gmail.com
Article Publishing History
Received: 19/04/2018
Accepted After Revision: 02/06/2018
Among different abiotic stresses (cold, temperature, salinity, drought, oxidative stress etc.) moisture stress is the most important limiting factor for crop production and is becoming an increasingly severe problem in many regions of the world. The aim of the current study is to identify some keys genes that are responsible for drought tolerant related traits, in the selected genotypes of Little millet (BL-8, MM-23, MM-10, BL-15-1, RLM-37, OLM-203, BL-4, JK-8). Genotypes belonging to diverse genetic background were grown under stress and control conditions for the identification of moisture stress tolerant traits. A set of known moisture stress related gene orthologs were selected for expression analysis using semi quantitative RT-PCR analysis. Expression analysis of these drought responsive gene orthologs (Amino-transferase, Thionin-osthi, Aquaporin, Synaptotagmin, CDPK, Scythe protein, Ta NAC-2, Ec NAC-67, NAM, U2-SnRNP, Hv NAC, Os NAC-29-2, MPK 17-1, DQP1, DQP 2, DQP 3, DQP 4, DQP 6 ) had given a differential expression under moisture stress as compared to controlled traits. Majority of these genes were up-regulated in the genotypes RLM-37, MM-23, MM-10, BL-4, BL-8 and BL-15-1 under moisture stress condition and these findings was found to be in correlation with the estimated biochemical traits (Proline, Chlorophyll, Carbohydrate and Protein). This can be taken as a base for drought tolerance response of the crop, which may be useful for further validation studies of the candidate genes for drought tolerance in the millet species as well as other crop plants.
Abiotic Stress, Drought, Semi Quantitative Rt-Pcr
Sushmitha B, Arun H P, Dubey M, Chandel G. Transcript Analysis of the Known Moisture Stress Responsive Gene Orthologs Among Different Genotypes of Little Millet, Panicum Sumatrense. Biosc.Biotech.Res.Comm. 2018;11(2).
Sushmitha B, Arun H P, Dubey M, Chandel G. Transcript Analysis of the Known Moisture Stress Responsive Gene Orthologs Among Different Genotypes of Little Millet, Panicum Sumatrense. Biosc.Biotech.Res.Comm. 2018;11(2). Available from: https://bit.ly/30bI0ql
Introduction
The exceptional tolerance of millets toward diverse abiotic stresses including drought, salinity, light and heat makes them a tractable system to study their stress responsive traits at the cellular, molecular and physiological levels (Bandyopadhyay et al., 2017). Several morpho-physiological and biochemical studies in millets have shown their stress adaptation strategies. Little millet is grown to a limited extent in India, up to altitudes of 2,100 m. It occurs wild in northern India and south East Asia. It belongs to the subfamily Panicoideae, tribe Paniceae, genus Panicum, species P. sumatrense, with chromosome number 36 (tetraploid) (Hiremath et al., 1990). Little millet is a domesticated form of the weedy species Panicum psilopodium (De Wet et al., 1983a). Introgression of genes between the two species is common (Hiremath et al., 1990). Little millet is comparable to other cereals in terms of fiber, fat, carbohydrates, and protein, and rich in phytochemicals including phenolic acids, flavonoids, tannins, and phytate (Pradeep and Guha 2011). Improved varieties of small millets could play a role in the “New Green Revolution”- a term coined to reflect novel strategies which will be required to deal with complex challenges in developing nations including increasing population and ever diminishing arable land. Like many other small millets, it is drought tolerant, pest and salt resistant,( Sivakumar et al., 2006b, Herder et al., 2010, Bhaskara and Panneerselvam 2013, Ajithkumar et al., 2014 Tang et al., 2017, Jaiswal et al., 2018).
Nagarjuna et al., (2016) have reported the identification and characterisation of an abiotic stress responsive protein kinase called CBL Interacting Protein Kinase (EcCIPK31-like) from drought tolerant crop, finger millet. Where, the upregulation was reported for first time under salinity, desiccation, oxidative and temperature stresses at seedling level in finger millet. The stress responsive nature of EcCIPK31-like to diverse stresses indicates that the gene could regulate multiple cellular tolerance traits and its further functional validation can highlight the relevance in abiotic stress. Similarly, it has been reported that Kodo millet is known to be highly drought and salt tolerant crop as ascertained by antioxidants and antioxidant enzymes levels. cDNA library was constructed from 6 days’ drought stressed seedlings. 5 ESTs differentially expressed under drought stress were characterized by DDRT-PCR and their expression profile was assessed by real time RT-PCR. Drought stress in Kodo millet led to the characterization of three up-regulated ESTs compared to two down-regulated, (Siddappa et al 2016).
Experimental results by Hittalmani, et al. (2017) revealed that, from whole genome sequencing and assembling process of ML-365 finger millet cultivar yielded 1196 Mb covering approximately 82% of total estimated genome size. Transcriptome analysis of low moisture stress and non-stress samples revealed the identification of several drought-induced candidate genes, which could be used in drought tolerance breeding. This genome sequencing effort had strengthened the plant breeders for allele discovery, genetic mapping, and identification of candidate genes for agronomically important traits.
In a study, physiological and transcriptomic comparisons between drought tolerant S. italica cultivar ‘Yugu1’ and drought-sensitive ‘An04’ were conducted by Tang, et al. (2017). They identified 20 candidate genes that contributed to germination and early seedling drought tolerance in S.italica. Finally their analysis provided a comprehensive picture of how different S.italica genotypes respond to drought, and may be used for the genetic improvement of drought tolerance in Poaceae crops.
Jaiswal, et al., (2018) reported de novo assembly-based transcriptomic signature of drought response induced by irrigation withdrawal in pearl millet. They found 19,983 differentially expressed genes, 7,595 transcription factors, gene regulatory network having 45 hub genes controlling drought response. They also reported 34652 putative markers (4192 simple sequence repeats, 12111 SNPs and 6249 InDels). This Study had revealed the role of purine and tryptophan metabolism in ABA accumulation mediating abiotic response in which MAPK acts as major intracellular signal sensing drought.
The molecular biology of Little millet has been explored to a limited extent. Little millet is perhaps the least studied of the small millet species and there is much that requires investigation, including the establishment of a genetic map and sequenced genome. It is important to dissect the transcriptome information under stress condition for the identification and characterization of the key genes for moisture stress tolerance. The identified genes which were up-regulated under the moisture stress condition, can be taken as a base for drought tolerance response of the crop, which may be useful for further validation studies of the candidate genes for drought tolerance mechanism in the millet species as well as other crop plants.
Material and Methods
Sowing of Little millet (Panicum sumatrense) was done in trays. Moisture stress was imposed after 30 days of sowing, at the vegetative stage before panicle initiation for a set of eight Little millet genotypes under the controlled environmental conditions as shown in figure 1. Temperature was maintained around 30±2. Plants were watered normally once in a day before the stress imposition and the leaf samples are harvested when the soil moisture content in the stress trays as reached below 10%. The harvested samples were stored immediately in liquid nitrogen for RNA isolation. The RWC was calculated based on the formula suggested by Barr and Weatherley (1962) as follows:
 |
Figure 1: Plant morphology of Eight Little millet Genotypes (RLM-37, OLM-203, MM-10, MM-23, JK-8, BL-4, BL-8, BL-15-1) under Control and Stress condition at Vegetative stage before Panicle Initiation |
![]()
Where FW = fresh weights of leaf taken immediately after excision TW = Turgid weight of leaf DW = Dry weight of leaf dried at 70 °C for 48 h. Leaf carbohydrate content was estimated by phenol sulphuric acid method proposed by Krishnaveni et al., (1984). The total carbohydrate present in the sample solution was calculated as given below using the standard graph. Absorbance corresponds to 8 ml of the test = ‘x’ mg of glucose. 100 ml of the sample solution contains = x / Sample volume X 10 mg of glucose. Leaf proline content was estimated by Acid ninhydrin method described by Bates et al., (1973). Free proline content in the sample was estimated by referring to a standard curve made from known concentrations of Proline by taking following
formula:

Leaf Chlorophyll content was estimated by acetone method developed by Arnon (1949). The amount of Chlorophyll present in leaf sample was calculated by using following equation:
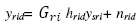
The values were expressed in milligram per gram of fresh weight. Where, A = Absorbance at specific wavelength, V = final volume of chlorophyll extract in 80% acetone, Wt = fresh weight of tissue extract. Leaf Protein content was estimated as per the method given by Lowry et al., (1951). From the standard graph the amount of protein in the unknown solution was calculated. The amount of protein present in the unknown solution is mg (μg of protein). The effect of moisture stress under stress and control condition in genotypes of Little millet was analysed statistically by calculating factorial CRD using OP-STAT, an online computerized software developed at BHU. RNA was isolated using TRIzol (Invitrogen, USA) and the concentration was determined using Nanodrop spectrophotometer ND-1000® (Nanodrop technologies USA). cDNA was prepared by using BIORAD iScript cDNA synthesis kit as per manufacturer’s instructions.
Semi-Quantitative RT-PCR reactions were carried out in 20 µl of the solutions using gene specific primers and Actin gene primer as internal control. The reaction was performed by adding following components in order into the PCR tubes for amplification: cDNA of 2.0 µl, 10X PCR buffer of 2.0 µl, (2Mm) dNTP of mix 1.0 µl, Primer Forward of 1.0 µl, Primer Reverse of 1.0 µl, (5U/ul) Taq polymerase of 0.2 µl, Nanopure water 1,500 ng/µl of 11.8 µl. Amplifications were performed by a cycles of 2 min at 950C followed by 35 cycles each of 15 sec at 950C, 30 sec at 56-620C, and 30 sec at 680C and final extention of 1 min at 720C.
 |
Figure 3a |
 |
Figure 3b |
Separation of amplified fragments was carried out using Bio-Rad gel electrophoresis assembly. PCR amplification products were analyzed by Agarose gel electrophoresis on 1.5% agarose gel stained with Ethidium Bromide solution (0.5 μg/ml). The gel was run in 1X TBE buffer at 70-80 Volts for 45 minutes to 1.5 h. Standard ladders of 100bp size were used. The resultant PCR product was then resolved on 1.5% Agarose gel followed by digitalization of fluorescence data to numerical values using GelQuant.NET Analyze. The relative expression of genes was expressed in terms of fold change (Increase/Decrease) under water stress with respect to their
control.
Results and Discussion
Wide variation for Relative water content values was recorded in stress tissue compared to control one’s among all tested Little millet genotypes. RWC of leaf samples ranged from 14.711% to 67.9% in stress plant leaf tissues and 67.821% to 95.073% in control plant leaf tissues. The drought tolerant Little millet genotype OLM-203 has the highest RWC value (67.9%) in stress tissue followed by MM-23 (64.83%), JK-8 (64.255%), RLM-37 (64.028%), MM-10 (52.966%), BL-4(35.48%). whereas, susceptible genotypes BL-15-1 (14.711%), BL-8 (14.194%) showed the minimum RWC in stress tissues. Two Little Millet genotypes RLM-37 and OLM-203 had shown lower decrease in RWC with values of 0.141% and 13.85% respectively as given in table 1. This clearly indicates that the two genotypes have the ability to retain more water during moisture stress which helps in sustaining the metabolic and physiological activities of plants. A wide variation for proline content was recorded in stress tissue compared to that of control one’s for eight diverse Little millet genotypes (BL-8, MM-23, MM-10, BL-15-1, RLM-37, OLM-203, BL-4, JK-8). The proline content ranged from 0.192 to 7.869 µ mole/tissue under stress; whereas under control condition proline content ranged from 0.015 to 0.204 µmole/tissue. A significant increase in proline has been observed in response to water stress, favouring osmotic adjustment. When comparing fold increase in proline content under stress over control among eight genotypes BL-15-1 was found with (63.460) higher fold increase followed by BL-8 (52.789), MM-10 (42.200), BL-4 (13.264), MM-23 (9.121), JK-8 (3.514), OLM-203 (1.670) and RLM-37 (1.343) as given in table 1. Enhanced proline accumulation in leaf tissues of plants exposed to water stress condition is considered as one of the major trait for the phenotypic characterization of plants for abiotic stress tolerance (Zhu et al., 2006).
| Table 1: Percentage change (Fold Increase or Decrease) in the RWC, Proline, Chlorophyll, Carbohydrate and Protein content for Eight Genotypes of Little millet (BL-8, MM-23, MM-10, BL-15-1, RLM-37, OLM-203, BL-4, JK-8) under Control and Stress condition. | |||||
| Genotype | Decrease in RWC % | Fold increase proline | Fold decrease
Total Chlorophyll |
Fold increase Carbohydrate | Fold increase Protein |
| RLM-37 | 13.85 | 1.343 | 1.042 | 1.274 | 1.481 |
| OLM-203 | 0.141 | 1.670 | 1.229 | 1.522 | 1.821 |
| JK-8 | 27.275 | 3.514 | 1.334 | 1.502 | 2.875 |
| MM-23 | 28.702 | 9.121 | 1.221 | 2.202 | 1.431 |
| MM-10 | 14.855 | 42.200 | 2.011 | 1.633 | 3.604 |
| BL-4 | 39.63 | 13.264 | 1.583 | 1.671 | 8.746 |
| BL-8 | 56.82 | 52.789 | 1.368 | 1.043 | 8.457 |
| BL-15-1 | 80.362 | 63.460 | 1.463 | 2.705 | 1.750 |
A wide variation for chlorophyll content was recorded in stress tissues for eight Little millet genotypes (BL-8, MM-23, MM-10, BL-15-1, RLM-37, OLM-203, BL-4, JK-8). Chlorophyll a, Chlorophyll b and Total Chlorophyll content ranged from 0.783 to 2.441 mg/tissue, 0.403 to 1.332 mg/tissue, 1.330 to 3.811 mg/tissue respectively for stress leaf tissue where as under control condition it ranged from 1.223 to 3.075 mg/tissue, 0.597 to 3.006 mg/tissue, 1.819 to 6.047 mg/tissue respectively. The genotype MM-10 had the highest fold decrease of 2.011 mg/tissue followed by BL-4 (1.583), BL-15-1 (1.463), BL-8 (1.368), JK-8 (1.334), OLM-203 (1.229), MM-23 (1.221) RLM-37 (1.042) in the total chlorophyll content, Where as in case of chlorophyll a, the genotype MM-23 showed highest fold decrease of 2.00 followed by BL-4 (1.749), BL-15-1 (1.641), MM-10 (1.619), JK-8 (1.571), BL-8 (1.319), OLM-203 (1.208), RLM-37 (1.016) and in chlorophyll b, the genotype JK-8 showed the highest fold decrease of 3.00 followed by MM-10 (2.665), BL-8 (1.481), MM-23 (1.360), BL-4 (1.314), OLM-203 (1.270), BL-15-1 (1.159), RLM-37 (1.149) as given in the table 1.
The carbohydrate content ranges from 234.221 to 612.222 mg/tissue under stress condition whereas 153.907 to 302.313 mg/tissue under control condition. BL-15-1 (2.705) had the highest fold increase followed by MM-23 (2.202), BL-4 (1.671), MM-10 (1.633), OLM-203 (1.522), JK-8 (1.502), RLM-37 (1.274), and BL-8 (1.043) as given in table 1. The wide variation for protein content was recorded in stress tissues for eight Little millet genotypes (BL-8, MM-23, MM-10, BL-15-1, RLM-37, OLM-203, BL-4, JK-8). The protein content ranged from 0.040 to 0.586 mg/tissue under stress condition, whereas 0.027 to 0.080 mg/tissue under control condition. BL-4 (8.746) had the highest fold increase followed by BL-8 (8.457), MM-10 (3.604), JK-8 (2.875), OLM-204 (1.821), BL-15-1 (1.750), RLM-37 (1.481), MM-23 (1.431) as given in table 1.
Expression pattern of drought stress responsive genes in little millet genotypes under moisture stress condition
Semi quantitative RT-PCR was performed to analyze the expression pattern of eighteen differentially expressed transcripts in Little millet under moisture stress versus control condition. The genes include Amino-transferase, Thionin-osthi, Aquaporin, Synaptotagmin, CDPK, Scythe protein, Ta NAC-2, Ec NAC-67, NAM, U2-Sn RNP, Hv NAC, Os NAC-29-2, MPK 17-1, DQP-1, DQP-2, DQP-3, DQP-4, DQP-6. Semi quantitative RT-PCR analysis showed differential expression of these eighteen transcripts in Little millet genotypes under stress with respect to the control condition. The results are discussed below in detail. Note: The increase or decrease in the Fold value was calculated my measuring the band intensity and size using GelQuantNET software.
| Table 2: List of Eighteen Genes, with their Primer sequences, GenBank Acc. No./Locus ID’s and Annealing temperatures used for Expression Analysis for Eight Genotypes of Little millet (BL-8, MM-23, MM-10, BL-15-1, RLM-37, OLM-203, BL-4, JK-8) under Control and Stress condition. | |||
| Gene | GenBank Acc. No./ Locus ID | Forward and Reverse sequence (5’-3’) | Tm |
| Amino Transferase | LOC_Os08g39300 | CTGAGTGGAGTGGAGATGGT
GTTCGTTGTGCTTCAGATCC |
610C |
| Thionin osthi | GT090938 | TCAACGCTGCTCTGGGAAAT
GGCTTGGTCGCAACTCTCAA |
580C |
| Synaptotagmin | GT090932 | TCTTGCAAGGTGCCAAATCTG GGCTGTGGCGTCCACTTAA | 580C |
| U2 Sn RNP | GT090867 | TGTGACCGACTTCCGTGAAG CCACGGTTGCAGCTGTTCT | 590C |
| Scythe protein | GT090877 | CCAGACACTAGCAGCACACATG CATCCCTTGCTCTGTTTGCA | 590C |
| Aquaporin | GT090849 | CCCGTTCAAGAGCAGGTCTTA CCTGTTTGGACTGGCATCTCA | 610C |
| CDPK | GT090918 | CAGAATTGACAGAGAATGAAATCCGATGGTTCCGCTGTTGTCAATA | 580C |
| Os NAC 29-2 | NC_029266 | AAAGAAGGAGCAACGTGCATTCTTGTGGATTCTGCACAGC | 560C |
| MPK 17-1 | GT090884 | TGTCGATGGATTGTCTGAAAAAGT TGCCGCGGTCTTTGGA | 560C |
| Ta NAC-2 | JN621240 | GATTTGGTCGGGATTTCAGA GCTCCATCATCGTCTCCTCT | 570C |
| Ec NAC-67 | KU500625 | CACTGCAAAGGAGGAGGAAG CTTCTTGGGCACCATCATCT | 580C |
| Hv NAC | JX855805 | CTACGACGACATCCAGAGCA
GTCATCCATTCCGCTTCTGT |
580C |
| NAM | LOC_Os03g21060 | CAAGACCAACTGGATCATGC
TTCTTGTAGATCCGGCACAG |
620C |
| DQP1 | LOC_Os08g36920 | AGTACATGATCCGATTCGAC
GTCCTGTAGCCGGAGATGAC |
65.40C |
| DQP 2 | LOC_Os11g26760 | GTGAAGGAGGAGCACAAGAC
TTGATCTTCTCCTTGATTCC |
640C |
| DQP 3 | LOC_Os01g44390 | CGATGTCGGTGAGCTCGT
GGTCTCGATGCGCTTGAC |
63.50C |
| DQP 4 | LOC_Os03g20550 | ATCAATCACACCATCTAGGC
GTATCTGGGGAAATTACAGTTG |
610C |
| DQP 6 | LOC_Os04g49550 | GAGCTAGAGAGGAAGACGATG
ATGATGACGATGTCCCTGTC |
64.10C |
You J., Hu H., Xiong L.(2012) have confirmed that OsOAT is a direct target gene of the stress-responsive NAC transcription factor SNAC2 (Li et al., 2008). In addition, OsOAT over expressing plants show significantly increased tolerance to oxidative stress mainly through enhancing ROS-scavenging capacity and pre-accumulation (You et al., 2012). The RT-PCR of Amino-transferase showed up-regulation in the transcript level by 2.25, 5.65 fold in the genotypes OLM-203, MM-23 respectively as shown in graph 1. The transcriptional analysis revealed that PvOAT was strongly induced by drought stress and it has also been reported that the expression of PvOAT was higher in leaves than that in root and stem of common bean (Phaseolus vulgaris L.) by drought stress (Ji-bao et al., 2016). Thus the up-regulation of this transcript under water stress suggests that it may play a key role in identification of different transcription factors which are responsible for different drought tolerant mechanisms in Little millet.
Thionin Osthi belongs to oxidative stress category of genes. A report by Yamakawa et al. (2007) reveals that this gene showed 2.0 fold up-regulation under high temperature stress in Medicago truncatula. The RT-PCR of Thionin-osthi in this study showed up-regulation in the transcript level by 10.15, 16.6 fold in the genotypes RLM-37, MM-23 respectively as shown in graph 1. Hence its induction in moisture stress in Little millet may be attributed due to the presence of the cis acting elements, suggesting an important role of this gene in combating oxidative stress.
Aquaporin belongs to major intrinsic protein super family which functions as a membrane channel. Over expression of a Panax ginseng tonoplast, aquaporin enhances drought and salt tolerance ability in transgenic Arabidopsis plants (Yanhui et al., 2007). But the RT-PCR of Aquaporin showed a negligible up-regulation in the transcript level by 1.16, 1.26, 1.42 fold in the genotypes BL-8, MM-23, OLM-203 respectively as shown in graph 1.
It has been shown that Synaptotagmin imparts calcium dependent freezing tolerance via membrane resealing and also loss of function of this gene reduces cell viability and plasma membrane integrity in Arabidopsis (Yamazaki et al., 2008). RT-PCR of Synaptotagmins showed an up-regulation in the transcript level by 2.92 fold in the genotype JK-8 as shown in graph 2. Identification and up-regulation of Synaptotagmin under moisture stress indicates its role in stress signal transduction and tolerance which needs to be further elucidated.
Calcium-dependent protein kinases play important role in signalling pathways for various stress responses (Ray et al., 2007, Sheen J. 1996). The RT-PCR of CDPK showed up-regulation in the transcript level by 1.21, 5.25 fold in the genotypes MM-10, BL-4 respectively as shown in graph 2. In several previous studies, induction and expression of CDPK(s) have been reported to be higher in tolerant cultivars in different abiotic stresses (Kawasaki et al., 2001, Li et al., 2008). The CIPK family of 26 protein kinases regulates the function of several ion transporters near the cell membrane to restore ion homeostasis under stress situations (Chaves-Sanjuan et al., 2014).
The differentially induced expression of OsCIPK genes by different stresses and the examples of improved stress tolerance of the OsCIPK transgenic rice suggest that rice CIPK genes have diverse roles in different stress responses and some of them may possess potential usefulness in stress tolerance improvement of rice (Xiang et al., 2007). Thus, suggesting its putative role in cell signalling pathway and in combating drought stress.
Scythe protein has been observed as a novel reaper-binding apoptotic regulator in vertebrates. Research by Thress et al., suggests that the Scythe protein might work by regulating the folding and activity of the molecules that make up the signaling pathway that controls apoptosis (Thress et al., 1998). The RT-PCR of Scythe protein showed up-regulation in the transcript level by 2.01, 29.20 fold in the genotypes MM-23 and MM-10 respectively and a negligible level of up-regulation was found in BL-4 by 1.31 fold as shown in graph 2.NAC (NAM, ATAF, and CUC) is a plant specific gene family of transcription factors. A few NAC genes from Arabidopsis and Brassica have been reported to be responsive in various environmental stresses (Shao et al., 2015). Over expression of various NAC genes have also been reported to significantly improve drought tolerance in transgenic rice Shao et al., (2015).
RT-PCR of NAM showed a negligible up-regulation in the transcript level by 1.07, 1.07, 1.20 fold in the genotyes BL-8, BL-15-1, MM-23 respectively, Where as a significant up-regulation was found in the genotype OLM-203 by 3.75 fold as shown in graph 3. The RT-PCR analysis of the up-regulation of this transcript under moisture stress suggests that it may play an important role in the cross-linking of different signalling pathways in Little millet.
In wheat, NAC TFs are known to be involved in processes such as senescence and nutrient remobilization as well as responses to abiotic and biotic stresses, ranging from stripe rust to abiotic stresses including drought and salt tolerance (Xia et al., 2010a). Out of four genes Ta NAC-2, Ec NAC-67, Hv NAC, Os NAC-29-2, The RT-PCR analysis of Ta NAC-2 showed up-regulation in the transcript level by 2.35, 8.66 fold in the genotypes MM-10, MM-23 respectively and RT-PCR analysis of Ec NAC-67 showed up-regulation in the transcript level by 4.44, 10.76 fold in the genotypes BL-15-1, MM-23 respectively as shown in graph 3. The RT-PCR analysis of Hv NAC showed negligible up-regulation in the transcript level by 1.30, 1.75 in the genotypes RLM-37, BL-4 respectively and RT-PCR analysis of Os NAC-29-2 showed a significant up-regulation in the transcript level by 13.99 fold in the genotype RLM-37 as shown in graph 4.
Alternative splicing takes place in highly specialized structures within nucleus called spliceosomes consisting of five small nuclear ribonucleoprotein particles, SnRNPs (U1, U2, U4/6, and U5) and other non-SnRNPs (Reddy ASN. 2001). RT-PCR of U2-SnRNP showed an up-regulation in the transcript level by 3.12, 4.65 fold in the genotypes MM-23, RLM-37 respectively shown in graph 4. Similar gene induction of U2-SnRNP has been reported among tolerant and susceptible cultivars of Foxtail millet (Charu Lata et al., 2010). These results suggest that U2-SnRNP may play a significant role in alternative splicing in Little millet and thus regulating gene expression.
MAP kinase signaling is one of the most important and conserved pathways in most cellular process as well as environmental stress responses (Lee et al., 2008, Moustafa et al., 2008). The RT-PCR analysis of MPK 17-1 showed an up-regulation in the transcript level by 7.99 fold in the genotype RLM-37 as shown in graph 5. MAP kinase gene has been reported to be induced due to dehydration, salinity and hyper-osmotic stresses (Moustafa et al., 2008). The activation of MPK 17-1 gene in moisture stress suggests that it may play an important role in the cross-linking of different signalling pathways to activate plant defense mechanisms in Little millet.
The AP2/EREBP genes play crucial roles in plant growth, development and biotic and abiotic stress responses and is one of the largest and specific transcription factor (TF) families in plants. Liu and Zhang have reported in G.hirsutum that, 151 non-repeated genes of the DREB and ERF subfamily genes were responsive to different stresses: 132 genes were induced by cold, 63 genes by drought and 94 genes by heat (Liu and Zhang 2017). The RT-PCR of DQP 1 showed an up-regulation in the transcript level by 2.78, 6.79 fold in the genotypes BL-15-1, BL-4 respectively as shown in graph 5.
Three of the four rice genes [(OsBIERF 1–4) Oryza sativa benzothiadiazole (BTH)-induced ethylene responsive transcriptional factors (ERF)] with a single conserved ERF domain were found to be up-regulated by salt, cold, drought, wounding as well as in an incompatible interaction between rice and fungal pathogen suggesting their role in biotic and abiotic stress (Jisha et al., 2015). In studies dealing with drought stress, Pelah et al. found a correlation between drought tolerance and accumulation of dehydrin proteins in Populus popularis (Pelah et al., 1997). The RT-PCR of DQP 2 showed an up-regulation in the transcript level by 2.23, 4.65 fold in the genotypes MM-23, BL-8 respectively. There was a negligible up-regulation found in the genotypes JK-8, MM-10, OLM-203 by 1.37, 1.37, 1.47 fold respectively as shown in graph 5.
A total of 44.67% and 47.21% MYB genes were found up and down-regulated in Arabidopsis under cold stress, respectively in the case of drought stress, many MYB genes have been isolated and demonstrated to be involved in drought responses in plants (Mmadi et al., 2017). The transcriptional activation of cuticular wax biosynthesis by MYB96 contributed to drought resistance in Arabidopsis thaliana (Seo et al., 2011). The RT-PCR of DQP 3 showed a significant up-regulation in the transcript level by 5.15 fold in BL-8 genotype as shown in graph 6. Altogether, these evidences demonstrated the versatility and importance of this gene family in plants.
Members of the large family of WRKY transcription factors are involved in a wide range of developmental and physiological processes, most particularly in the plant response to biotic and abiotic stress. RT-PCR analysis of Yu Y., Wang N., Hu R, Xiang F. showed that in whole soybean plant, 66 GmWRKYs exhibited distinct expression patterns in response to salt stress (Yu et al., 2016). The RT-PCR of DQP 4 showed a negligible up-regulation in the transcript level by 1.33, 1.40, 1.41 fold in the genotypes RLM-37, BL-8, MM-10 respectively and a significant level of up-regulation in the transcript was observed in the genotype BL-15-1 by 3.44 fold as shown in graph 6.
In a study by Liu K. et al. showed over-expression of OsCOIN protein, a RING finger protein in transgenic rice lines significantly enhanced their tolerance to cold, salt and drought, accompanied by an up-regulation of OsP5CS expression and an increase of cellular proline level (Liu et al., 2007). Salt and drought-induced RING FINGER1 (SDIR1), is involved in abscisic acid (ABA)-related stress signal transduction in Arabidopsis thaliana (Zhang et al., 2007). SDIR1 is expressed in all tissues of Arabidopsis and is up-regulated by drought and salt stress, but not by ABA (Zhang et al., 2007). The RT-PCR of DQP 6 showed an up-regulation in the transcript level by 2.61 fold in the genotype OLM-203 and a negligible level of up-regulation was noticed in the genotype MM-23 by 1.11 fold as shown in graph 6.
Out of 18 transcripts under control and stress condition for eight genotypes of Little millet, a significant level of up-regulation was observed among the following:
- Genotype MM-23 showed a higher level of up-regulation for the genes Aminotransferase, Ta NAC-2 Ec NAC-67 and Thionin osthi by 5.65, 8.66, 10.76, 16.60 fold respectively.
- Genotype RLM-37 showed a significant up-regulation for the genes U2 Sn RNP, MPK-17-1, Thionin osthi and Os NAC-29-2 by 4.65, 7.99, 10.15 and 13.99 fold respectively
- Genotype BL-4 showed an up-regulation in the transcript level by 5.25 and 6.79 fold for the genes CDPK and DQP1 respectively.
- Genotype MM-10 showed a greater level of up-regulation for the gene scythe protein by 29.20 fold.
- Genotype BL-8 showed an up-regulation for the genes DQP 2 and DQP 3 by 4.65 and 5.15 fold respectively.
- BL-15-1 genotype showed a significant up-regulation for the transcript Ec NAC-67 by 4.44 fold.
Conclusion
Current study helps us to identify the key genes expressed in response to moisture stress in the selected Little millet genotypes. Induction of transcripts Amino-transferase, Thionin-osthi, Aquaporin,Synaptotagmin, CDPK, Scythe protein, Ta NAC-2, Ec NAC-67, NAM, U2-Sn RNP, Hv NAC, Os NAC-29-2, MPK 17-1, DQP1, DQP 2, DQP 3, DQP 4, and DQP 6 suggests that these genes may impart drought avoidance capacity to the tolerant genotypes. Genes which were up-regulated suggests their function in positive regulation in adaptation of the moisture stress under the drought condition, this can be taken as a base for drought tolerance response of the crop, which may be useful for further validation studies of the candidate genes for drought tolerance in the millet species as well as other crop plants.
Acknowledgments
Seed material was provided by ZARS, Jagdalpur, and KVK Shivpuri. Department of Biotechnology, Ministry of Science and Technology, Govt. of India are thankfully acknowledged for providing the financial support.
References
Ajithkumar I.P., Panneerselvam R., (April 2014) ROS scavenging system, osmotic maintenance, pigment and growth status of Panicum sumatrense roth. Under drought stress. Cell Biochem Biophys. Vol. 68, No 3: Pages 587-95 doi:10.1007/s12013-013-9746-x.
Bandyopadhyay T, Muthamilarasan M, Prasad M. (2017) Millets for Next Generation Climate-Smart Agriculture. Frontiers in Plant Science Vol. 8: Pages 1266. doi:10.3389/fpls.2017.01266.
Bhaskara J., Panneerselvam R. (2013) Accelerated reactive oxygen scavenging system and membrane integrity of two Panicum species varying in salt tolerance. Cell Biochem Biophys. Vol. 67, No 3: Pages 885-92.
Chaves, M.M., Maroco J.P., Pereira J.S. (Jan 2003) Understanding plant responses to drought-from genes to the whole plant. Functional Plant Biology Vol. 30: Pages 239-264 doi: 10.1071/FP02076
Charu Lata, Sahu P.P., Prasad M. (Mar 2010) Comparative transcriptome analysis of differentially expressed genes in foxtail millet (Setaria italica L.) during dehydration stress. Biochemical and Biophysical Research Communications Vol. 393 No 4: Pages 720–727 doi: 10.1016/j.bbrc.2010.02.068.
Chaves-Sanjuan A. et al. (2014) Structural basis of the regulatory mechanism of the plant CIPK family of protein kinases controlling ion homeostasis and abiotic stress. Proc. Natl. Acad. Sci. USA Vol. 111, No 42: Pages 4532–4541.
Den Herder G., Van Isterdael G., Beeckman T., De Smet I. (Nov 2010) The roots of a new green revolution. Trends Plant Sci. Vol 15, No 11: Pages 600-7.
De Wet J. M. J., Prasada Rao K. E., Brink D. E., (1983a). Systematics and domestication of Panicum sumatrense (Graminae). J. d’agriculture Tradit. Bot. appliquée Vol. 30, No 2: Pages 159–168.
Hiremath S. C. Patil G. N. V. Salimath S. S. (1990). Genome homology and origin of Panicum sumatrense (Gramineae). Cytologia (Tokyo) Volume 55 No 2: Pages 315-319 doi: 10.1508/ cytologia.55.315.
Ji-bao CHEN, Yuan-nan CAO, Zhao-yuan ZHANG, Shu-min WANG, Jing WU, Lan-fen WANG (2016). Cloning of the OAT gene and the correlation between its expression and drought tolerance in Phaseolus vulgaris L. Journal of Integrative Agriculture Vol. 15, No 5: Pages 973-982 doi:10.1016/S2095-3119(15)61283-7.
Jisha V., Dampanaboina L., Vadassery J., Mithöfer A., Kappara S., Ramanan R. (2015) Overexpression of an AP2/ERF Type Transcription Factor OsEREBP1 Confers Biotic and Abiotic Stress Tolerance in Rice. PLOS ONE Vol. 10, No 6: Page e0127831 doi:10.1371/journal.pone.0127831.
Kawasaki S., Borchert C., Deyholos M., et al., ( April 2001) Gene expression profiles during the initial phase of salt stress in rice, Plant Cell Vol. 13, No 4: Pages 889–905.
Lee J.S., Huh K.W., Bhargava A., Ellis B.E. (Dec 2008) Comprehensive analysis of protein– protein interactions between Arabidopsis MAPKs and MAPK kinases help define potential MAPK signaling modules. Plant Signaling Behav. Vol. 3 No 12: Pages 1037–1041.
Li A., Wang X., Leseberg CH., Jia J., Mao L. (2008) Biotic and abiotic stress responses through calcium-dependent protein kinase (CDPK) signaling in wheat (Triticum aestivum L.) Plant Signaling Behav. Vol. 3 No 9: Pages 654–656.
Liu C., Zhang T. (Jan 2017) Expansion and stress responses of the AP2/EREBP superfamily in cotton. BMC Genomics Vol. 18 No 1:Page 118.
Liu K., Wang L., Xu Y., Chen N., Ma Q., Li F., Chong K. (Sep 2007) Overexpression of OsCOIN, a putative cold inducible zinc finger protein, increased tolerance to chilling, salt and drought, and enhanced proline level in rice. Planta Vol. 226, No 4: Pages 1007-16. doi: 10.1007/s00425-007-0548-5.
Mmadi M.A., Dossa K., Wang L., et al., (Dec 2017) Functional Characterization of the Versatile MYB Gene Family Uncovered Their Important Roles in Plant Development and Responses to Drought and Waterlogging in Sesame. Genes.Vol. 8 No 12: Page E362. doi:10.3390/genes8120362.
Moustafa K., Vos D.L., Leprince A., Savoure A., Lauriere C. (2008) Analysis of the Arabidopsis Mitogen-activated protein kinase families: organ specificity and transcriptional regulation upon water stresses. Scholarly Research Exchange Vol. 2008 No 2008 12 pages Article ID 143656 doi:10.3814/2008/143656.
Nagarjuna, K. N., Parvathi, M. S., Sajeevan, R. S., Pruthvi, V., Mamrutha, H. M., Nataraja, K. N. (2016). Full-length cloning and characterization of abiotic stress responsive CIPK31-like gene from Finger millet, a drought-tolerant crop. Current Science, Vol.111 No 5: Pages 43-45.
Nisha Chhabra and Amarjeet Kaur (Part A 2017) Studies on physical and engineering characteristics of maize, pearl millet and soybean. Journal of Pharmacognosy and Phytochemistry Vol. 6 No 6: Pages 01-05.
Pelah D., Wang W., Altman A., Shoseyov O., Bartels D. (1997) Differential accumulation of water stress related proteins, sucrose synthase and soluble sugars in Populus species that differnin their water stress response. Physiol Plant Vol. 99 No 1: Pages 153–159 doi:10.1111/j.1399-3054.
Pradeep SR., Guha M., (Jun15 2011) Effect of processing methods on the nutraceutical and antioxidant properties of Little millet (Panicum sumatrense) extracts. Food Chem. Vol. 126, No 4: Pages 1643-7
Ray S., Agarwal P., Arora R., et al., (2007) Expression analysis of calcium-dependent protein kinase gene family during reproductive development and abiotic stress conditions in rice (Oryza sativa L. ssp. indica), Mol. Genet. Genomics Vol. 278, No 5: Pages 493–505 DOI: 10.1007/s00438-007-0267-4.
Reddy ASN. (2001) Nuclear pre mRNA splicing in plants, Crit. Rev. Plant Sci. Vol. 20 No 6: Pages 523–571 doi:abs/10.1080/20013591099272.
Sarika Jaiswal, Tushar, J., Antala, Mandavia, M.K., Meenu Chopra, Rahul Singh Jasrotia, Rukam S. Tomar, Jashminkumar Kheni, Angadi, U.B., Iquebal, M.A., Golakia, B.A., Anil Rai and Dinesh Kumar (2018). Transcriptomic signature of drought response in pearl millet (Pennisetum glaucum (L.) and development of web-genomic resources. Nature, 8: Article number-3382.
Seo P.J., Lee S.B., Suh M.C., Park M.J., Go Y.S., Park C.M. (2011) The MYB96 Transcription Factor Regulates Cuticular Wax Biosynthesis under Drought Conditions in Arabidopsis. The Plant Cell Vol.23 No 3: Pages 1138-1152.
Shao H., Wang H., Tang X. (2015) NAC transcription factors in plant multiple abiotic stress responses: progress and prospects. Frontiers in Plant Science Vol. 6: Page 902. doi:10.3389/fpls.2015.00902.
Sha Tang, Lin Li, Yongqiang Wang, Qiannan Chen, Wenying Zhang, Guanqing Jia, Hui Zhi, Baohua Zhao & Xianmin Diao. (2017). Genotype-specific physiological and transcriptomic responses to drought stress in Setaria italica (an emerging model for Panicoideae grasses) Nature, 7: Article number-10009.
Shailaja Hittalmani, Mahesh, H.B., Meghana Deepak Shirke, Hanamareddy Biradar, Mohanrao, A. (2017). Genome and Transcriptome sequence of Finger millet (Eleusine coracana (L.) Gaertn.) provides insights into drought tolerance and nutraceutical properties. BMC Genomics, 18 : 465.
Sheen J. (1996) Ca2+-dependent protein kinases and stress signal transduction in plants, Science Vol. 274, No 5294: Pages 1900–1902.
Shinozaki, K., Yamaguchi-Shinozaki K., Seki M. (Oct 2003). Regulatory network of gene expression in the drought and cold stress responses. Curr Opin Plant Biol Vol. 6 No 5: Pages 410-417.
Siddappa, N., Raghu ,G. K., and Devaraj Varadahally, R. (2016). Identification of Drought-Responsive Transcripts in Kodo Millet (Paspalum scrobiculatum L.) International Journal of Innovative Research & Development, Vol.5 No 11: Pages 140-143.
Sivakumar S., Mohan M., Franco O. L., Thayumanavan B., (2006b). Inhibition of insect pest á-amylases by little and finger millet inhibitors. Pestic. Biochem. Physiol. Vol.85: Pages 155–160 doi:10.1016/j.pestbp.2005.11.008
Thress K., Henzel W., Shillinglaw W., Kornbluth S. (1998) Scythe: a novel reaper-binding apoptotic regulator. The EMBO Journal Vol. 17 No 21: Pages 6135-6143. doi:10.1093/emboj/17.21.6135.
Xia N., Zhang G., Liu X.-Y., Deng L., Cai G.-L., et al., (2010a) Characterization of a novel wheat NAC transcription factor gene involved in defense response against stripe rust pathogen infection and abiotic stresses. Mol. Biol. Rep. Vol. 37 No 8: Pages 3703–3712 doi: 10.1007/s11033-010-0023-4.
Xiang Y., Huang Y., Xiong L. (Jul 2007) Characterization of stress responsive CIPK genes in rice for stress tolerance improvement. Plant Physiol. Vol. 144 No 3: Pages 1416–1428 DOI: https://doi.org/10.1104/pp.107.101295.
Xiong, L., Schumaker K.S., Zhu J.-K. (2002). Cell signaling during cold, drought, and salt stress. The plant cell Vol. 14: S165-S183.
Yamaguchi-Shinozaki, K., Shinozaki K. (2006) Transcriptional regulatory networks in cellular responses and tolerance to dehydration and cold stresses. Annual Review of Plant Biology Vol. 57: Pages 781-803 doi:10.1146/annurev.arplant.57.032905.105444.
Yamazaki T., Kawamura Y., Minami A., Uemura M (2008) Calcium-dependent freezing tolerance in Arabidopsis involves membrane resealing via synaptotagmin SYT1, Plant Cell Vol. 20 No 12: Pages 3389–3404, DOI:10.1105/tpc.108.062679.
Yanhui Peng, Wuling Lin, Weiming Cai, Rajeev Arora (2007) Overexpression of a Panax ginseng tonoplast aquaporin alters salt tolerance, drought tolerance and cold acclimation ability in transgenic Arabidopsis plants, Planta Vol. 226, No 3: Pages 729-40 DOI 10.1007/s00425-007-0520-4.
You J., Hu H., Xiong L. (2012) An ornithine ä-aminotransferase gene OsOAT confers drought and oxidative stress tolerance in rice. Plant Science Vol 197: Pages 59–69, doi: 10.1016/j.plantsci.2012.09.002.
Yu Y., Wang N., Hu R., Xiang F. (Jun 2016) Genome-wide identification of soybean WRKY transcription factors in response to salt stress. Springer Plus Vol 5 No 1: Page 920. doi:10.1186/s40064-016-2647-x.
Zhang Y., Yang C., Li Y., et al. (2007) SDIR1 Is a RING Finger E3 Ligase That Positively Regulates Stress-Responsive Abscisic Acid Signaling in Arabidopsis. The Plant Cell Vol 19 No 6:Pages 1912-1929 doi:10.1105/tpc.106.048488.
Zhu, H., Ding G.H., Fang K., Zhao F.G., and Qin P. (2006) New perspective on the mechanism of alleviating salt stress by spermidine in barley seedlings. Plant Growth Regulation Vol 49 No 2-3: Pages 147-156 doi:10.1104/pp.107.105882.