Department of Plant Pathology, Tamil Nadu Agricultural University, Coimbatore, India-641003
Corresponding author Email: pn40785@gmail.com
Article Publishing History
Received: 07/04/2019
Accepted After Revision: 06/06/2019
Bacterial leaf blight pathogen, Xanthomonas oryzae pv. oryzae is a major biotic constraint in rice production which eventually make the global food grain sustainability under stress. Host resistance being the only strategy of management of the disease, the present study focused on screening of 28 near isogenic lines and/or varieties under field condition which revealed resistant reaction of the following lines viz., IRBB 21, IRBB 57, IRBB 58 and IRBB 61 against Xanthomonas oryzae pv. oryzae (NCBI Accession No. MH464904). Besides, R genes viz., Xa21, xa5 and xa13 were identified in these near isogenic lines where different combinations of genes have been identified. With the intention of understanding the kind of avr genes present in X. o. pv. oryzae isolate and thereby elucidating the kind of interaction persists among rice and bacterial leaf blight pathogen, gene specific amplification and identification of avr genes were performed. The results revealed that avr genes viz., PthXo1, avrxa3, AvrXa10, Avr/pthC8b and Avr/pth56B were positively amplified whereas Avr/pth3 was absent in the isolate even at repeated attempts. Interaction of X. o. pv. oryzae against ADT 38 rice variety through SDS PAGE analysis of the pathogen and abiotic agents (Biotin (0.1mM), Riboflavin (0.5mM), Chloramphenicol (0.1mM), Ethrel (1µl/ml)) applied samples showed that defense related proteins were induced alike in all the samples treated irrespective of the treatment when compared to untreated control.
Avr gene Bacterial leaf blight, R gene, Rice
Praveen N. M, Monisha S,Ramanathan S. Studies on Interaction of Rice and Bacterial Leaf Blight Causing Xanthomonas Oryzae Pv. Oryzae. Biosc.Biotech.Res.Comm. 2019;12(2).
Praveen N. M, Monisha S,Ramanathan S. Studies on Interaction of Rice and Bacterial Leaf Blight Causing Xanthomonas Oryzae Pv. Oryzae. Biosc.Biotech.Res.Comm. 2019;12(2). Available from: https://bit.ly/2L3lyer
Copyright © Praveen et al., This is an open access article distributed under the terms of the Creative Commons Attribution License (CC-BY) https://creativecommns.org/licenses/by/4.0/, which permits unrestricted use distribution and reproduction in any medium, provide the original author and source are credited.
Introduction
Plants are confronted consistently with pathogens and based on the molecular determinants in plants (Pathogen recognition receptor (PRRs), Resistance genes) and in pathogen (Pathogen associated molecular patterns (PAMPs), Avr genes) both interacts to build defense responses where PRRs and PAMPs interacts to provide broad spectrum resistance (horizontal resistance) whereas R genes and Avr genes interaction may result in gene specific resistance (vertical resistance) (Vander Plank, 1984). Rice is one of the most significant cultivated food crops which feeds half of the population worldwide where the demand for rice is progressively increasing in developing countries (Khush and Jena, 2009) whereas bacterial leaf blight pathogen, Xanthomonas oryxae pv. oryzae (Xoo) becoming one among the important delimiting biotic factor that reduces rice production up to 81 per cent (Kumar et al. 2012). Host resistance is the only management strategy that could strongly dependent to contain the disease as the practice of application of chemicals in management of disease fronting adverse backlashes (Rajpurohit et al. 2011, Dokku et al. 2013, Suh et al. 2013; Sundaram et al. 2014; Arunakumari et al. 2016; Hajira et al. 2016; Gao et al. 2018).
Hence, most favorable strategies for crop improvement program for the disease resistance is either selection of donor source of resistance against Xoo and thereby using for resistance breeding or by introduction of resistance genes into desired variety or cultivars. Since understanding of interaction between rice and bacterial leaf blight pathogen is still remain ambiguous, researches on elucidating the mechanism of interaction and defense response has significant role. Interaction between rice and bacterial leaf blight pathogen is tend to follow the classical gene-for-gene relationship (Flor 1971), however, there exists distinctive differences for the interaction of R genes of bacterial leaf blight disease from characterized R genes of other crops where researchers also identified closely linked molecular markers (Yoshimura et al. 1995, Sonti 1998; Rao et al. 2002, Gu et al. 2008; Khan et al. 2015; Arunakumari et al. 2016; Hajira et al. 2016; Chukwu et al. 2019).
Exploration, identification, and utilization of new resistant germplasms in rice breeding are the strategical steps to control the bacterial blight disease of rice. The Xa21 gene has been successfully introgressed into several elite rice varieties and hybrid rice parental lines all over the world either singly or in combination with other major resistance genes such as Xa4, xa5, and xa13 (Singh et al. 2001; Joseph et al. 2004; Sundaram et al. 2008; Sundaram et al. 2009; Perumalsamy et al. 2010; Pandey et al. 2013).
Most of the reported R genes of other crop-pathogen systems are dominant in nature, unlike in rice where one-third of the R-genes conferring resistance to bacterial leaf blight disease have been reported as recessive (Verdier et al. 2012). Forty resistant genes, Xa1 to Xa39 have been identified in rice system which were against bacterial leaf blight pathogen (Kim et al. 2015).
Such R genes correspondingly recognize Avr gene products of the bacterial leaf blight pathogen directly or indirectly which could mediate defense response. However, such responses to contain the disease never ensue often in a host-pathogen system as the pathogen could evolve and evoke the virulence to cause the disease with distinct avirulence and virulence factor. Hence, screening of rice lines against various strains or pathotypes of bacterial leaf blight pathogen has prominent role to play. TAL effector dependent R genes induces downstream expression of R genes, for example, Xa10 contains a binding element for the TAL effector, AvrXa10 (EBEAvrXa10) in its promoter, and which induces Xa10 expression (Tian et al. 2014). Hence, Avr and R gene plays imperative changes in host system to combat bacterial leaf blight disease in rice. Therefore, identification of the presence of Avr genes in the pathotypes prevailing in each location and its virulence nature against the cultivars and Near Isogenic Lines (NILs) could help in developing resistant donor parent for the crop improvement program through resistant breeding. Moreover, for successful deployment of stable resistance genes, their characterization and availability of tightly linked markers will greatly facilitate the development of new versions of cultivars (Vikal and Bhattia, 2017).
The present study is focused on identification of R genes present in NILs which confer downstream defense response after recognition (directly or indirectly) of Avr genes and also the marker assisted genotyping of major R genes of bacterial leaf blight of rice.
Material and Methods
Bacterial leaf blight symptom of rice was isolated from various locations viz., Coimbatore, Aduthurai, Gudalur, Krishnagiri, Vellur, Theni, Wayanad, Hyderabad and New Delhi during the survey and were brought into laboratory for isolation of the pathogen under in vitro condition. Briefly, under in vitro, the diseased samples collected were sliced off into leaf bits containing both the healthy and infected portions were transferred into an Eppendorf tube containing sterile water. Later, the leaf bits were crushed using sterile rod to release bacterial colonies as ooze and the loopful of suspension was streaked onto the autoclaved, solidified Peptone sucrose agar media (CaNO3-0.5g, FeSO4. 7H2O- 0.5g, sucrose-15g, peptone-5g, Na2HPO4. 7H2O-2g, agar-15g, distilled water-1000ml) poured into the Petri dish. Triplicates of samples were maintained for isolation and were incubated at 25oC for the formation of yellow, mucoid, doom shaped colonies with entire margins were developed and were sub cultured for further studies. Pathogenicity of the isolate was proved in bacterial leaf blight susceptible varieties viz., ADT 38 and TN1. Seeds of the susceptible varieties were collected in a jute bag and immersed overnight in water and on next day, the soaked seeds in jute bag were kept overnight, air tightened inside the hay to sprout. Sprouted seeds were collected on the next day for sowing in pots filled with clay loamy soil. Seedlings emerged from the pots were transplanted to another one after 15 days of sowing which were maintained in a growth chamber of temperature of 250C and 85-90% relative humidity. Forty eight hours old bacterial suspension prepared in nutrient broth conditioned to 0.1 value in spectrophotometer at 600nm were employed for testing pathogenicity nature of the isolate through clip inoculation method (Kauffman et al. 1973).
Concisely, clip inoculation method was performed into the seedlings with maximum tillering stage where surface sterilized scissors were used to nick out the top 5cm of leaves of the seedling after dipping into the bacterial suspension prepared earlier in the nutrient broth. Remaining bacterial suspension was sprayed onto the cut ends and margins of leaves to permeate the entry of bacterial colonies into the leaves and cause the infection. Observation for symptom appearance from the day of inoculation was recorded and the organism was re-isolated from the lesion and compared with that of the original isolate and hence proved the pathogenicity. Characterization of bacterial leaf blight pathogen was instigated by isolation of total genomic DNA from the bacteria which was performed using lysis buffer method (Chen and Kuo, 1993) where the bacterial culture was multiplied in 100 ml nutrient broth kept for 48hr in rotary shaker at 180rpm. Saturated culture was harvested in 1.5 ml of Eppendorf tube and allowed for centrifugation for 3 min at 12,000 rpm. 200 μl of lysis buffer (40 mM Tris-acetate pH 7.8, 20 mM sodium-acetate, 1 mM EDTA, 1% SDS) was suspended in the cell pellet and lysed by vigorous pipetting. 66 µl of 5M NaCl was added and mixed well which remove most proteins and cell debris and then the viscous mixture was centrifuged at 12,000 rpm for 10 min at 4°C.
Clear supernatant obtained after centrifugation was transferred into new vial and an equal volume of chloroform was added followed by vortex until the solution turned milky. Subsequently, centrifuged the solution at 12,000 rpm for 3 minutes, and the supernatant extracted was transferred to another vial and the DNA was precipitated with 100% ethanol, washed twice with 70% ethanol, dried, and redissolved in 50 μl of 1 x TE buffer. Later, the quality of the DNA was assessed by 1.2% agarose gel electrophoresis. To assess the genetic identity of the pathogen, PCR was performed using 16s rRNA primers (27F: 5′-AGAGTTTGATCCTGGCTCAG-3′) and 1492R (5′-GGTTACCTTGTTACGACTT-3′) which shows the amplicon size of 1537bp. The PCR product was outsourced for sequencing to identify upto the species level of bacteria. Hence, the bacteria identified as Xanthomonas oryzae pv. oryzae (Xoo) through molecular tools were used for screening of bacterial leaf blight pathogen against rice cultures in field. Thirty one rice lines were sown in rice beds and were transplanted to fields after 20 days of sowing such that each lines had a minimum of ten hills in a row. Integrated nutrient management practices were followed as per the package of practices for rice crop. Maximum tillering was obtained at 45 days old crop and in which clip inoculation was carried out following the method of Kauffman et al. (1973) described above. As the bacterial infection spread from the cut ends of the leaves, the lesion length was recorded at 7 and 14 days after inoculation of bacterial suspension and the lesion length was compared with that of the resistance reaction to bacterial leaf blight mentioned in Standard Evaluation System of Rice.
Screening of these culture lines were carried out twice during the period of 2017-18 and 2018-19 and confirmed the resistant reaction nature of the culture lines in comparison with the existing susceptible cultivars. To understand the interaction between host and pathogen, presence of resistance genes and avr genes in rice cultures and X. o. pv. oryzae respectively were identified using polymerase chain reaction amplification through the gene specific primers. Genomic DNA of thirty one rice lines were isolated using conventional CTAB method in which 0.5g of leaf samples were ground with 500µl of CTAB buffer using pestle and mortar. The contents were transferred to a vial and kept in water bath at 65oC for 15 minutes followed by cooled at room temperature. Chloroform and Isoamyl alcohol mixture at 24:1 ratio were prepared and equal volume of the solution was added into vial and centrifuged at 12,000 rpm for 15 minutes. Aqueous phase thus formed was transferred into a new vial and poured equal volume of ice cold isopropanol and incubated overnight at -20oC. Pellet was separated from the solution by centrifugation at 12,000rpm for 15 minutes and added 200µl of 70% of ethanol. Air dried the pellet and kept the pellet dissolved in sterile water at -20oC for further studies.
Genomic DNA in each rice lines were checked at 0.8% of agarose gel electrophoresis. A total of 25µl of reaction mixture for polymerase chain reaction amplification of genomic DNA of 31 rice lines were performed. Primers viz., xa5 (F: 5’ GCACTGCAACCATCAATGAATC 3’; R: 5’ CCTAGGAGAAACTAGCCGTCCA 3’), xa13 (F: 5’ CCTGATATGTGAGGTAGT 3’); R: 5’ GAGAAAGGCTTAAGTGC 3’) and PTA 248 (F: 5’ AGACGCGGGAAGGGTGGTTCCCGGA 3’); (R: 5’ AGACGCGGGTAATCGAAAGATGAAA 3’) (Robert et al. 1992) each at 100pm/µl were used to identify the presence of R genes (xa5, xa13 and Xa21).
Initial denaturation of 5 min at 94°C followed by 40 cycles of 1 min of denaturation at 94°C, 1 min of annealing at 55°C for xa5 and xa13 gene primers and 59oC for Xa21 gene primer, and 1 min of extension at 72°C and final extension was 10 min at 72°C. PCR amplification of genomic DNA of X. o. pv. oryzae was performed using Avr gene primers specific to avrxa3 gene, Avr/pth3 gene, Avr/pth56B, Avr/pthC8b and AvrXa10, Pth/Xo1 (Table 4.). PCR conditions followed were initial denaturation of 5 minutes at 94°C followed by 40 cycles of 1 minute of denaturation at 94°C, 1 minute of annealing at 55°C and 1 minute of extension at 72°C and final extension was 10 minutes at 72°C. Resistance genes in host and avr genes in the pathogen are the important molecular determinants that downstream the defense mechanism in host system by induction of secretion of several proteins.
An experiment was laid out to detect the presence of proteins secreted after bacterial leaf blight susceptible rice variety, ADT 38 was confronted with the pathogen and abiotic agents viz., Biotin (0.1mM), Riboflavin (0.5mM), Chloramphenicol (0.1mM), Ethrel (1µl/ml). Virulent bacterial colonies were inoculated into nutrient broth and incubated for 48hr in BOD incubator with 180rpm till the spectrophotometer read 0.1 at 600nm. Bacterial suspension was clip inoculated into the 40 days old plant followed by abiotic agents were applied 24 hr after the inoculation. Leaves were collected 24hr after application of abiotic agents separately from each and were subjected to protein extraction for SDS PAGE analysis to detect the presence of protein fractions secreted.
Results and Discussion
Organism causing bacterial leaf blight disease in rice was isolated from the leaf samples collected during the survey in Coimbatore, Aduthurai, Gudalur, Krishnagiri, Vellur, Theni, Wayanad, Hyderabad and New Delhi under in vitro condition. The virulent strains of the bacterial leaf blight isolates were demarcated with that of the non-virulent one as the former appeared yellow, round, mucoid, convex colonies after 72 hr of incubation whereas the latter remained white, slimy, mucoid and convex as detailed by Webster and Gunnell (1992). Pathogenicity tests for 9 isolates were proved for all the 9 isolates in ADT 38 and TN1 which showed initial symptom of the disease after 7 days of inoculation. Virulence spectrum of X. o. pv. oryzae isolates were compared after inoculating in ADT 38 and TN1 varieties revealed that Coimbatore isolate imparted highest virulence than any other. While comparing the lesion size formed by each isolate, highest lesion size was observed in the variety inoculated with Coimbatore isolate whereas the least was observed with New Delhi isolate.
Genomic DNA of nine bacterial leaf blight pathogens were extracted and the molecular characterization of the nucleic acids using 16S rRNA primers showed amplification at 1200 bp and partial sequencing of the isolates revealed the identity of the organism as Xanthomonas oryzae pv. oryzae. Screening of 29 rice lines along with improved samba mahsuri (resistant check) and TN1 (Susceptible check) against bacterial leaf blight pathogen (Coimbatore) showed formation of typical bacterial leaf blight symptom after 7 days of inoculation. Lesion size were recorded during 7th and 14th day of inoculation, revealed the resistant reaction offered against the pathogen by the rice cultures. When compared to the resistant check, improved samba mahsuri, rice cultures showed better resistive potential. Three rice cultures viz., IRBB 57, IRBB 58, IRBB 61 were categorized as resistant among the 29 culture lines according to the Standard Rice Evaluation System of IRRI, Philippines whereas 11 were moderately resistant (IRBB 7, IRBB 8, IRBB 11, IRBB 13, IRBB 14, IRBB 21, IRBB 50, IRBB 56, IRBB 63, IRBB 64 and IRBB 66) and 13 culture lines (IRBB 1, IRBB 3, IRBB 4, IRBB 5, IRBB 10, IRBB 51, IRBB 52, IRBB 53, IRBB 54, IRBB 55, IRBB 59, IRBB 60 and IRBB 62) were moderately resistant (Table 1, Table 2).
Table 1: Field screening of rice cultures against bacterial leaf blight pathogen during 2017-18
| Sl. No | Rice lines | Average lesion size (cm)
7th day |
Average lesion size (cm) 14th day | Resistant reaction |
| 1. | IRBB 1 | 7.5l | 8.2ijk | MS |
| 2. | IRBB 3 | 5.6f | 6.6efgh | MS |
| 3. | IRBB 4 | 7.8l | 8.2ijk | MS |
| 4. | IRBB 5 | 6.8jk | 7.0ghi | MS |
| 5. | IRBB 7 | 3.9c | 4.2bc | MR |
| 6. | IRBB 8 | 4.8d | 5.0cde | MR |
| 7. | IRBB 10 | 5.1e | 5.5def | MR |
| 8. | IRBB 11 | 6.1hi | 6.6efgh | MS |
| 9. | IRBB 13 | 3.6c | 4.2bc | MR |
| 10. | IRBB 14 | 3.6c | 4.3bc | MR |
| 11. | IRBB 21 | 1.8a | 2.2a | R |
| 12. | IRBB 50 | 4.6d | 5.0cde | MR |
| 13. | IRBB 51 | 6gh | 6.6efgh | MS |
| 14. | IRBB 52 | 6.7ij | 7.0ghi | MS |
| 15. | IRBB 53 | 6.7ij | 7.0ghi | MS |
| 16. | IRBB 54 | 5.7fg | 6.6fghi | MS |
| 17. | IRBB 55 | 6.7ij | 7.0ghi | MS |
| 18. | IRBB 56 | 6.8k | 7.2ghif | MS |
| 19. | IRBB 57 | 2.6b | 2.9a | R |
| 20. | IRBB 58 | 2.5b | 2.8a | R |
| 21. | IRBB 59 | 6fg | 6.5efg | MS |
| 22. | IRBB 60 | 7k | 7.3hij | MS |
| 23. | IRBB 61 | 2.7b | 3.2ab | MR |
| 24. | IRBB 62 | 6.7ij | 7.5hij | MS |
| 25. | IRBB 63 | 3.7c | 4.3bcd | MR |
| 26. | IRBB 64 | 5.8fg | 6.6fghi | MS |
| 27. | IRBB 65 | 6fg | 6.7fghi | MS |
| 28. | IRBB 66 | 4.6d | 5.2cdef | MR |
| 29. | DV-85 | 2.8b | 3ab | MS |
| 30 | ISM | 5.9fg | 5.6def | MR |
| 31. | TN1 | 8m | 8.8jk | MS |
| CD (0.05) | 0.311 | 1.37 |
HR: Highly Resistant (<1cm); R: Resistant (1-3cm); MR: Moderately Resistant (3-6cm);
Moderately Susceptible (6-10cm); Susceptible (>10cm)
Table 2: Field screening of rice cultures against bacterial leaf blight pathogen during 2018-19
| Sl. No | Rice lines | Average lesion size (cm)
7th day |
Average lesion size (cm) 14th day | Resistant reaction |
| 1. | IRBB 1 | 6def | 8f | MS |
| 2. | IRBB 3 | 6def | 8f | MS |
| 3. | IRBB 4 | 6def | 8f | MS |
| 4. | IRBB 5 | 7f | 7f | MS |
| 5. | IRBB 7 | 2a | 4abc | MR |
| 6. | IRBB 8 | 5bcd | 5bcd | MR |
| 7. | IRBB 10 | 6def | 8f | MS |
| 8. | IRBB 11 | 4abc | 6def | MR |
| 9. | IRBB 13 | 4abc | 6def | MR |
| 10. | IRBB 14 | 4abc | 6def | MR |
| 11. | IRBB 21 | 2a | 4abc | MR |
| 12. | IRBB 50 | 5bcd | 6def | MR |
| 13. | IRBB 51 | 6def | 8f | MS |
| 14. | IRBB 52 | 7ef | 7f | MS |
| 15. | IRBB 53 | 7ef | 7f | MS |
| 16. | IRBB 54 | 6def | 8f | MS |
| 17. | IRBB 55 | 7ef | 7f | MS |
| 18. | IRBB 56 | 4abc | 6def | MR |
| 19. | IRBB 57 | 3ab | 3ab | R |
| 20. | IRBB 58 | 3ab | 3ab | R |
| 21. | IRBB 59 | 5bcd | 5bcd | MS |
| 22. | IRBB 60 | 7ef | 7ef | MS |
| 23. | IRBB 61 | 3ab | 3ab | R |
| 24. | IRBB 62 | 7.66ef | 7.66ef | MS |
| 25. | IRBB 63 | 4abc | 6def | MR |
| 26. | IRBB 64 | 4abc | 6def | MR |
| 27. | IRBB 65 | 6def | 6def | MR |
| 28. | IRBB 66 | 5bcd | 5bcd | MR |
| 29. | DV-85 | 3ab | 3ab | R |
| 30 | ISM | 5bcd | 5bcd | MR |
| 31. | TN1 | 8.8f | 7.66f | MS |
| CD (0.05) | 1.78 | 1.85 |
HR: Highly Resistant (<1cm); R: Resistant (1-3cm); MR: Moderately Resistant (3-6cm); Moderately Susceptible (6-10cm); Susceptible (>10cm)
Similarly, Bharathkumar et al. (2014) also tested the resistance reaction of rice and bacterial leaf blight pathogen and categorized pathotypes of X. o. pv. oryzae isolates prevailing in different states of India. Incongruence with the present study, Rajappan and Ravi (2015) evaluated twenty two gene pyramided rice cultures against bacterial leaf blight disease to estimate the resistant reactions against the disease during 2012-2015. These rice cultures were gene pyramided with resistance genes (Xa1 to Xa21) to bacterial leaf blight and with the combinations of the same which in turn give resistive nature to the lines. Identification of such combinations of R genes in the lines are advantageous for the identification of donor parent intended to development of a resistant varieties. Gene specific primers pertaining to xa5, xa13 and a functional marker of Xa21 gene (PTA 248) were amplified using rice genomes of twenty eight rice cultures and three cultivars where the results showed that polymorphic genotypes attained after gel electrophoresis were directly related to the resistant nature of the lines (Table 3).
Table 3: List of primers
| S.No. | Genes | Nucleotides | Annealing temperature |
| 1. | 16S rRNA | 27F: AGAGTTTGATCCTGGCTCAG
1492R: GGTTACCTTGTTACGACTT |
55 |
| 2. | PTA248 | F: AGACGCGGGAAGGGTGGTTCCCGGA
R: AGACGCGGGTAATCGAAAGATGAAA |
59 |
| 3. | xa13 | F: CCTGATATGTGAGGTAGT
R: GAGAAAGGCTTAAGTGC |
55 |
| 4. | xa5 | F: GCACTGCAACCATCAATGAATC
R: CCTAGGAGAAACTAGCCGTCCA |
55 |
| 5. | Avrxa3 | F: CATCTTGTTCCCACATCACG
F: GCCGGAATTGATCAGAAGAG |
55 |
| 6. | Avr/pth3 | F: AGGACATAATCAGGGCGTTG
R: CCAATACGGCGATTGACTCT |
55 |
| 7. | Avr/pth56b | F: GCCGGAATTGATCAGAAGAA
R: CATCTTGTTCCCACATCACG |
55 |
| 8. | Avr/pthC8b | F: GCCGGAATTGATCAGAAGAA
R: CATCTTGTTCCCACATCACG |
55 |
| 9. | AvrXa10 | F: ATCTGCTCCGTCAGTTCGAT
R: TGGCCTGTGTCCAACTGTAA |
55 |
| 10. | PthXo1 | F: GAGAGCATTGTTGCCCAGTT
R: CTGAAGTAGGGGACGGTTTG |
55 |
Gene specific amplification of Xa21 marker in 31 rice lines/varieties, 8 lines showed (IRBB 11, IRBB 55, IRBB 56, IRBB 57, IRBB 58, IRBB 59, IRBB 60, IRBB 61, IRBB 64, IRBB 65 and IRBB 66) amplification at 900 bp thereby confirmed the presence of Xa21 gene (Figure 1). Similarly, presence of xa5 gene amplification established after the comparison of gene amplification pattern similar to that of IRBB 5. The results revealed that IRBB 50, IRBB 59, IRBB 60, IRBB 61, IRBB 63, IRBB 64, IRBB 66 showed similar xa5 gene marker amplification at 270 bp (Figure 2). In the case of xa13 gene, rice lines viz., IRBB 51, IRBB 53, IRBB 54, IRBB 56, IRBB 58, IRBB 60, IRBB 61, IRBB 63, IRBB 65 were showed similar banding pattern of that of IRBB 13 which remarked the presence of xa13 gene (Figure 3).
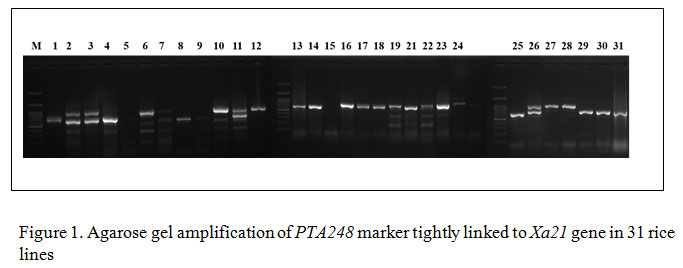 |
Figure 1: Agarose gel amplification of PTA248 marker tightly linked to Xa21 gene in 31 rice lines |
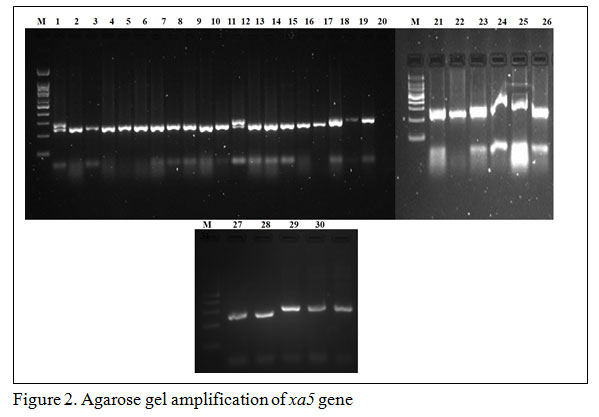 |
Figure 2: Agarose gel amplification of xa5 gene |
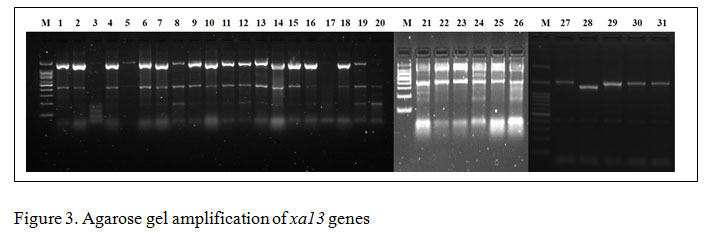 |
Figure 3: Agarose gel amplification of xa13 genes |
Genotyping of Xa21, xa5 and xa13 genes showed that all the three genes were present in IRBB 60 and IRBB 61. Xa21 and xa5 genes were present in IRBB 59 alone whereas xa5 and xa13 genes were amplified in IRBB 63 and IRBB 65 lines. To contain the bacterial leaf blight of rice, researchers paved major attention in introgression of multiple resistance genes through breeding program. Guvvala et al. (2013) have experimented gene pyramiding of R genes viz., Xa4, xa5, xa13 and Xa21 into bacterial leaf blight susceptible mahsuri variety and various such related works are in progress (Pinta et al. 2013, Pradhan et al. 2015, Chukwu et al. 2019).
Hence, the present study to identify the donor parents carrying R genes for resistance breeding program has pursued its importance. In the bacterial leaf blight pathogen, Xanthomonas oryzae pv. oryzae (NCBI Accession No. MH464904) after gene specific amplification of avr genes viz., pthXo1 (433 bp), avrxa3 (623 bp), AvrXa10 (562), Avr/pthC8b (420 bp) and Avr/pth56B (623) were obtained whereas Avr/pth3 gene was absent in the strain (Figure 4). As a testimonial to the above results, Wu et al. (2007) came into conclusion that avrXa3 gene containing three nuclear localization signal (NLS) motifs which is consistently present in all members of the avrBs3/pthA family was identified in JXOIII strain of X. o. pv. oryzae. Numbers of the avr/pth genes vary among different strains of X. o. pv. oryzae.
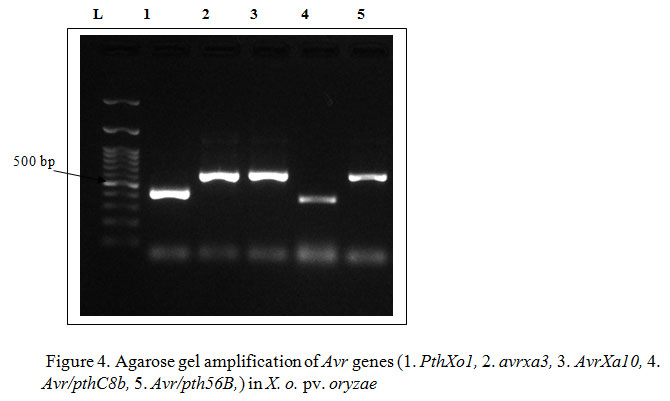 |
Figure 4: Agarose gel amplification of Avr genes (1. PthXo1, 2. avrxa3, 3. AvrXa10, 4. Avr/pthC8b, 5. Avr/pth56B,) in X. o. pv. oryzae |
Similar to the identification of avr genes in individual strain, researchers have identified presence of 15 avr3/pth genes in Korean race 1 (KACC10331) (Lee et al. 2005) whereas Yang and White (2004) demonstrated 25–32 avr/pth genes in different Philippine strains. After inoculation of X. o. pv. oryzae suspension into rice lines carrying R genes, avr gene products interact correspondingly with that of R gene product and based on the downstream action of the corresponding gene interaction compatibility or incompatibility were established to cause disease susceptibility or resistance. Disease susceptibility gene, Os8N3 or susceptible allele of recessive R gene xa13 were found to be specifically induced by TAL effectors, PthXo1 (Romer et al. 2010, Yin et al. 2017).
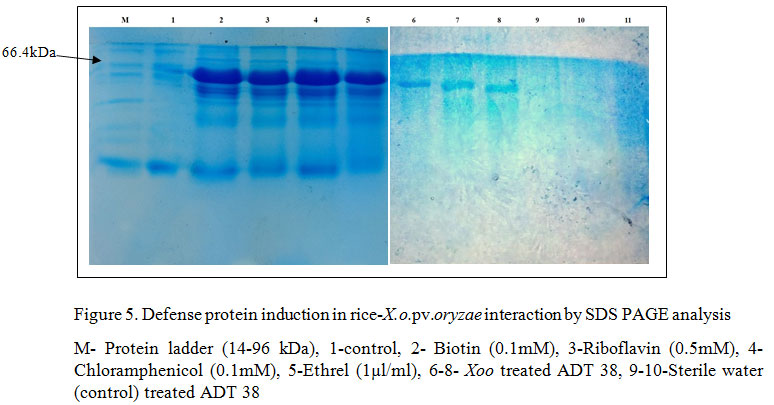
Hence, as PthXo1 gene was present in the Coimbatore strain of X. o. pv. oryzae, it could induce the expression of disease susceptibility allele, Xa 13 gene which thereby results in susceptibility of the line. Avr/pth56B and Avr/pthC8b were also identified in X. o. pv. oryzae isolate of Coimbatore, whereas there are ambiguity over the detailed evidence regarding the interaction with the R genes and those avr genes. In an extension to the identification type of R genes and avr genes present in the host and pathogen, to analyze the PR protein or other defense protein induction, SDS PAGE analysis was performed in rice variety, ADT 38, after inoculation of the pathogen followed by abiotic chemical agents (Biotin (0.1mM), Riboflavin (0.5mM), Chloramphenicol (0.1mM), Ethrel (1µl/ml)). The results revealed that comparing to the untreated control, all the treatments has induced the defense proteins consistently at 66.4 kDa thereby demonstrated that the susceptible rice variety could induce defense proteins with the application of abiotic agents (Figure 5). SERK2 protein is a 69 kDa proteins which is secreted as immune response mediated by the LRR receptor kinases, Xa21 and the bacterial pathogen interaction. Hence, the apparent molecular weight of the protein observed in SDS-PAGE are consistent with that of SERK2 protein. In congruence to the above study, Wu et al. (2011) has conducted an experiment to compare the expression of PR proteins in compatible and incompatible interactions and results showed that in both the case the induction was noticed. Six out of ten PR proteins (PR1, PR2, PR3, PR4b, PR8, and PR-pha) showed enhanced expression in Xa21-mediated resistance responses at late stages after inoculation with X. o. pv. oryzae (Hou et al. 2011).
Conclusion
Interaction between rice and bacterial leaf blight pathogen, Xanthomonas oryzae pv. oryzae resulted in induction of defense related proteins which were observed through SDS PAGE analysis. The defense related proteins synthesized were mediated by the direct and/or indirect interaction of resistance genes and corresponding avirulence genes of rice cultures and X. o. pv. oryzae respectively. PthXo1, avrxa3, AvrXa10, Avr/pthC8b and Avr/pth56B were the Avr genes identified from X. o. pv. oryzae, Coimbatore isolate.
Acknowledgements
We acknowledge the help and cooperation of Department of Plant Pathology, Centre for Plant Protection Studies, Tamil Nadu Agricultural University, Coimbatore, India. Department of Rice, Centre for Plant Breeding and Genetics, Tamil Nadu Agricultural University, Coimbatore, India
Conflict of Interest
All authors have declared no conflicts of interest in this communication
References
Arunakumari, K., Durgarani, C. V., Satturu, V., Sarikonda, K. R., Chittoor, P. D. R., Vutukuri, B., Laha, G. S., Nelli, A. P. K., Gattu, S., Jamal, M., Prasadbabu, A., Hajira, S. and Sundaram, M. (2016): Marker-Assisted Pyramiding of Genes Conferring Resistance Against Bacterial Blight and Blast Diseases into Indian Rice Variety MTU1010. Rice Science Vol 23(6) Pages 306-316
Bharathkumar, S., Gnanamanickam, S. S., Jitendra, K., Archana, B., Yasin, B. S. K., Amit, D. and Deepak, K. N (2014): Culture disease reaction of rice pathogen Xanthomonas oryzae oryzae prevailing in India on rice cultivars. The Bioscan Vol. 9, Pages 1257-1262.
Chen, W. P. and Kuo, T. T. (1993): A simple and rapid method for the preparation of Gram negative bacterial genomic DNA. Nucleic Acids Research Vol. 21 No. 9 Pages 2260-2264.
Chukwu, S. C., Rafii, M. Y., Ramlee, S. I., Ismail, S. I., Oladosue, Y., Okporie, E., Onyishi, G., Utobo, E., Ekwud, L., Swaray, S. and Jalloh, M. (2019): Marker-assisted selection and gene pyramiding for resistance to bacterial leaf blight disease of rice (Oryza sativa). Biotechnology Biotechnological Equipment, DOI: 10.1080/13102818.2019.1584054.
Dokku, P., Das, K. M. and Rao, G. J. N. (2013): Pyramiding of four resistance genes of bacterial blight in Tapaswini, an elite rice cultivar, through marker-assisted selection. Euphytica Vol. 192 Pages 87-96.
Flor, H. H. (1971): Current status of the gene-for-gene concept. Annual Review of Phytopathology Vol. 9 Pages 275-296.
Gao, L., Fang, Z., Zhou, J., Li, L., Lu, L., Li, L., Li, T., Chen, L., Zhang, W., Zhai, W. and Peng, H. (2018): Transcriptional insights into the pyramided resistance to rice bacterial blight. Scientific Reports Vol. 8 Pages 12358
Gu, K., Sangha, J. S., Li, Y. and Yin Z (2008): High resolution genetic mapping of bacterial blight resistance gene Xa10. Theoretical and Applied Genetics Vol. 116 Pages 155-163.
Guvvala, L. D., Koradi, P., Shenoy, V. and Marella, L. S. (2013): Making an Indian traditional rice variety Mahsuri, bacterial blight resistant using marker-assisted selection. Journal of Crop Sciences and Biotechnology Vol. 16 Pages 111-121.
Hajira, S. K., Sundaram, R. M., Laha, G. S., Yugander, A., Balachander, S. M., Viraktamath, B. C., Sujatha, K., Balachiranjeevi, C. H. Pranathi, K., Anila, M., Bhaskar, S., Abhilash, V., Mahadevaswamy, H. K., Kousik, M., Dilip kumar, T., Harika, G. Rekha, G. (2016): A Single-Tube, functional marker-based multiplex PCR assay for simultaneous detection of major bacterial blight resistance genes Xa21, xa13 and xa5 in rice. Rice Science Vol 23(3) Pages 144−151.
Hou, M., Xu, W., Bai, H., Liu, Y., Li, L., Liu, L., Liu, B. and Liu., G. 2011. Characteristic expression of rice pathogenesis-related proteins in rice leaves during interactions with Xanthomonas oryzae oryzae. Plant Cell Reports Vol 31 No 5 Pages 895-904
Joseph, M., Gopalakrishnan, S., Sharma, R. K., Singh, V. P., Singh, A. K., Singh, N. K. and Mohapatra, T. (2004): Combining bacterial blight resistance and basmati quality characteristics by phenotypic and molecular marker assisted selection in rice. Molecular Breeding. Vol. 13 pages 377–387.
Kauffman, H. E., Reddy, A. P. K., Hsieh, S. P. Y., Merca, S. D. (1973): An improved technique for evaluating resistance of rice varieties to Xanthomonas oryzae. Plant Disease Reports Vol. 57: 537–541.
Khan, M. W., Abbasi, F. M., Masood, M. S., Rabbani, A., Abbasi, m. F., Sajid, M., Khan, U., Ahmad, H. (2015): Identification of bacterial blight resistance gene Xa7 in rice (Oryzae sativa L.) through STS marker. International Journal of Biosciences Vol 6 Pages 318-32
Khush, G. and Jena, K. K. (2009): Current status and future prospects for research on blast resistance in rice (Oryza sativa). In: Wang GL, Valent B (eds) Advances in genetics, genomics and control of rice blast disease. Springer, New York, pp 1-10.
Kumar, P. N., Sujatha, K., Laha, G. S., Rao, K. S., Mishra, B., Viraktamath, B. C., Hari, Y., Reddy, C. S., Balachandran, S. M., Ram, T., Madhav, M. S., Rani, N. S., Neeraja, C. N., Reddy, G. A., Shaik, H. and Sundaram, R. M. (2012): Identification and fine mapping of Xa33, a novel gene for resistance to Xanthomonas oryzae pv. oryzae. Phytopathology Vol. 102 Pages 222-228.
Lee, B. M., Park, Y. J., Park, D. S., Kang, H. W., Kim, J. G., Song, E. S., Park, I. C., Yoon, U. H., Hahn, J. H., Koo, B. S., Lee, G. B., Kim, H., Park, H. S., Yoon, K. O., Kim, J. H., Jung, C. H., Koh, N. H., Seo, J. S. and Go, S. J. (2005): The genome sequence of Xanthomonas oryzae pathovar oryzae KACC10331, the bacterial blight pathogen of rice. Nucleic Acids Research. 33, 577-86.
Pandey, M. K., Rani, N. S., Sundaram, R. M., Laha, G. S., Madhav, M. S, Srinivasa Rao, K., Sudharshan, I., Hari, Y., Varaprasad, G. S., Subba Rao, L. V., Suneetha, K., Sivaranjani, A. K. P. and Viraktamath, B. C. (2013): Improvement of two traditional Basmati rice varieties for bacterial blight resistance and plant stature through morphological and marker-assisted selection. Molecular Breeding.Vol 31 Pages 239–246.
Perumalsamy, S., Bharani, M., Sudha, M., Nagarajan, P., Arul, L., Saraswathi, R., Balasubramanian, P., Ramalingam, J. (2010): Functional marker-assisted selection for bacterial leaf blight resistance genes in rice (Oryza sativa). Plant Breeding. Vol 129 Pages 400–406.
Pinta, W., Toojinda, T., Thummabenjapone, P. and Sanitchon, J. (2013): Pyramiding of blast and bacterial leaf blight resistance genes into rice cultivar RD6 using marker assisted selection. African Journal of Biotechnology Vol. 12 Pages 4432-4438.
Pradhan, S. K., Nayak, D. K., Mohanty, S., Behera, L., Barik, S. R., Pandit, E., Lenka, S. and Anandan, A. (2015): Pyramiding of three bacterial blight resistance genes for broad-spectrum resistance in deepwater rice variety, Jalmagna. Rice Vol. 8. No. 19 Pages 1-14.
Rajappan, K. and Ravi, V. (2015): Evaluation of gene pyramided rice cultures against bacterial leaf blight disease of rice. Journal of Rice Research Vol. 8 Pages 81-82.
Rajpurohit, D., Kumar, R., Kumar, M., Paul, P., Awasthi, A. A, Basha, P. O., Puri, A., Jhang, T., Singh, K., Dhaliwal, H. S. (2011): Pyramiding of two bacterial blight resistance and a semi dwarfing gene in Type 3 Basmati using marker-assisted selection. Euphytica Vol. 178 Pages 111–126.
Rao, K. K., Lakshminarasu, M., Jena, K. K. (2002): DNA markers and marker-assisted breeding for durable resistance to bacterial blight of rice. Biotechnology Advances. Vol. 20 No. 33-47.
Romer, P., Recht, S., Strau, T., Elsaesser, J., Schornack, S., Boch, J., Wang, S. and Lahaye, T. (2010): Promoter elements of rice susceptibility genes are bound and activated by specific TAL effectors from the bacterial blight pathogen, Xanthomonas oryzae pv. oryzae. New Phytologist Vol. 187 No. 1048–1057.
Singh, S., Sidhu, J. S., Huang, N., Vikal, Y., Li, Z., Brar, D. S., Dhaliwal, H. S., Khush, G. S. (2001): Pyramiding three bacterial blight resistance genes (xa5, xa13 and Xa21) using marker-assisted selection into indica rice cultivar PR106. Theoretical and Applied Genetics. Vol 102 Pages 1011–1015.
Sonti, R. V. (1998): Bacterial leaf blight of rice: new insights from molecular genetics. Current Science Vol. 74 Pages 206–212.
Suh, J. P., Jeung, J. U., Noh, T. H., Cho, Y. C., Park, S. H., Park, H. S., Shin, M. S., Kim, C. K., Jena, K. K. (2013): Development of breeding lines with three pyramided resistance genes that confer broad-spectrum bacterial blight resistance and their molecular analysis in rice. Rice. Vol. 6 No. 5.
Sundaram, R. M., Vishnupriya, M. R., Biradar, S. K., Laha, G. S., Reddy, A. G., Rani, N. S., Sarma, N. P., Sonti, R. V. (2008): Marker assisted introgression of bacterial blight resistance in Samba Mahsuri, an elite indica rice variety. Euphytica.Vol 160 Pages 411–422.
Sundaram, R. M., Vishnupriya, M. R., Laha, G. S., Rani, N. S., Rao, P. S., Balachandran, S. M., Reddy, G. A., Sarma, N. P., Sonti, R. V. (2009): Introduction of bacterial blight resistance into Triguna, a high yielding, mid-early duration rice variety by molecular marker assisted breeding. Biotechnology Journal. Vol 4 Pages 400–407.
Sundaram, R. M. (2012): Identification and fine-mapping of Xa33, a novel gene for resistance to Xanthomonas oryzae pv. oryzae. Phytopathology Vol. 102 pages 222–228.
Sundaram, R. M., Chatterjee, S., Oliva, R., Laha, G. S., Leach, J. E., Sonti, R. V., Cruz, C. V. (2014): Update on bacterial blight of rice. Rice Vol 7(7) Pages 12.
Tian, D., Wang, J., Zheng, X., Gu, K., Qiu, C., Yang, X., Zhou, Z., Goh, M., Luo, Y., Murata-Hori, M., White, F. F., Yin, Z. (2014): The rice TAL effector-dependent resistance protein Xa10 triggers cell death and calcium depletion in the endoplasmic reticulum. The Plant Cell. Vol 26 Pages 497–515.
Vanderplank, J. E. (1984): Disease resistance in plants. 2nd Elsevier. 66-143p.
Verdier, V., Vera, C. C. and Leach, J. E. (2012): Controlling rice bacterial blight in Africa: needs and prospects. Journal of Biotechnology Vol. 159 Pages 320-328.
Vikal, Y. and Bhatia, D. (2017): Genetics and genomics of bacterial blight resistance in rice. Advances in international rice research. Pages 175-213.
Webster, R. K. and Gunnell, P. S. (1992): Compendium of rice diseases. The Disease compendium series of the American Phytopathological Society (USA).
Wu, Q., Hou, M. M., Li, L. Y., Liu, L. J., Hou, Y.X. and Liu, Y. X. (2011): Induction of pathogenesis-related proteins in rice bacterial blight resistant gene Xa21-mediated interactions with Xanthomonas oryzae oryzae. Journal of Plant Pathology Vol. 93 No. 2 Pages 455-459.
Wu, X. M., Li, Y. R., Zou, L. F., Chen, G. Y. (2007): Gene‐for‐gene relationships between rice and diverse avrBs3/pthA avirulence genes in Xanthomonas oryzae pv. oryzae. Plant Pathology Vol 56 Pages 26-34.
Yang, B. and White, F. (2004): Diverse members of the AvrBs3/PthA family of type III effectors are major virulence determinants in bacterial blight disease of rice. Molecular Plant-Microbe Interactions Vol. 17 Pages 1192-200.
Yin, Z. C., Gu, K. Y. and Tian, D. S. (2017): S. Patent No. 9,650,647. Washington, DC: U.S. Patent and Trademark Office.
Yoshimura, S., Yoshimura, A., Iwata, N., McCouch, S. R., Abenes, S. L., Baraoidan, M. R., Mew, T. W. and Nelson, R. J. (1995): Tagging and combining bacterial blight resistance genes in rice using RAPD and RFLP markers. Molecular Breeding Vol. 1 Pages 375–387.