1 School of Biotechnology, Devi Ahilya University Indore (M.P) India
2 Mata Gujri College of Professional Studies, Indore (M.P) India
Corresponding author email: sadhana0301@gmail.com
Orchid ID: https://orchid.org/0000-0001-8389-5919
Orchid ID: https://orcid.org/0000-0001-9967-0132
Article Publishing History
Received: 15/02/2023
Accepted After Revision: 22/03/2023
Cellobiose dehydrogenase (CDH) enzyme is secreted extracellularly by wood-rotting fungi of the phyla Basidiomycetes and Ascomycetes. The reducing ends of cellobiose, lactose, and maltose are oxidized by CDH to produce their respective lactones. These lactones are consequently converted into their carboxylic acids such as cellobionic acid, lactobionic acid, and maltobionic acid. Due to its commercial unavailability and its applications in various fields, there is a need for cost-effective CDH production. In the present work, Schizophyllum commune BCC26414 has been used for CDH production by solid-state fermentation (SSF).
CDH production was optimized by one factor at a time (OFAT) approach in terms of initial moisture content, inoculum size, incubation temperature, particle size, and fermentation time. BBD (Box-Behnken Design) was used to perform statistical optimization of CDH production using statistical software, Response Surface Methodology (RSM) Maximum CDH production was obtained when groundnut shell was used as a substrate at 30°C on 9th day of incubation, with 0.5mm to 1mm particle size, 2 ml inoculum size, and the initial moisture content 50% using Schizophyllum commune BCC26414. RSM enhances enzyme production to 1.6-fold as compared to unoptimized conditions. This is the first report on solid-state CDH production using groundnut shells as solid substrate. A variety of CDH applications have been reported in the fields of biomedical, biocatalysts, bioremediation, and biosensors. This study will be helpful in the cost-effective production of CDH for various applications.
Box-Behnken Design (BBD), Cellobiose dehydrogenase, Groundnut shell, Response Surface Methodology
(RSM), Schizophyllum commune BCC26414, Solid-state Fermentation (SSF).
Yadav V, Nighojkar S. Solid State Fermentation of Groundnut Shell by Schizophyllum commune BCC26414 for Production of Cellobiose Dehydrogenase. Biosc.Biotech.Res.Comm. 2023;16(1).
Yadav V, Nighojkar S. Solid State Fermentation of Groundnut Shell by Schizophyllum commune
BCC26414 for Production of Cellobiose Dehydrogenase. Biosc.Biotech.Res.Comm. 2023;16(1). Available from: <a href=”https://bit.ly/3xtKkeK“>https://bit.ly/3xtKkeK</a>
INTRODUCTION
Fungi are the most important and well-known group of microorganisms, having vast potential to degrade wood components such as cellulose, hemicellulose and lignin by the action of enzymes (Henriksson et al. 2000). One of these enzymes is Cellobiose dehydrogenase [E. C. 1.1.99.18], which is reported in several wood-decaying fungi belonging to the phylum basidiomycetes and ascomycetes (Henriksson et al. 2000; Banerjee et al. 2021). Cellobiose dehydrogenase (CDH) is mainly secreted by microbes including bacteria and fungi (Cameron et al., 2001). Disaccharides and oligosaccharides with β-1-4 glycosidic linkages are susceptible to oxidation of their reducing ends oxidized by CDH. These disaccharide and oligosaccharides produce their corresponding lactones and these lactones spontaneously convert to their carboxylic acids (Gangwar et al. 2021).
CDH is a monomeric protein and consist of two domains: a flavin domain that contains FAD and a heme domain that contains cytochrome b-type heme, which is connected by a serine and threonine-rich amino acid linker that is sensitive to protease. Catalytic and cellulose binding properties are present in FAD-containing fragment. Monosaccharides are the poor substrates for CDH due to lack of β-1-4 glycosidic linkages (Baminger et al. 2001; Gupta et al. 2014). CDH has important applications in many biotechnological fields such as pharmaceutical, cosmetics, food industries, and different clinical applications (Nyanhongo et al. 2017).
The solid-state fermentation (SSF) method can increase enzyme yield while minimizing the cost of enzyme production. The most frequent microorganisms utilized in SSF are filamentous fungi because they can grow on solid substrates with little water content (Mrudula and Murugammal, 2011). There are numerous studies describing the use of agro-industrial wastes as solid substrates in SSF, such as wheat straw (Li et al. 2008); while wheat bran, and rice- straw as substrates for submerged fermentation (Rai et al. 2020) for the production of CDH. SSF is reported to be especially suitable for fungi rather than bacteria (Patidar et al. 2018).
The SSF process is also known to be the most effective method for producing enzymes due to its high productivity, straightforward approach, inexpensive capital investment, low energy need, low water output, higher product recovery, and lack of foam buildup (Mrudula and Murugammal, 2011). In SSF, fungi primarily use agro-industrial wastes as a medium for the synthesis of metabolites and enzymes. Fungi are more appropriate for use in SSF processes since they can grow and exploit agricultural waste as their natural environment. Fungi have been believed to be the organisms most adapted to SSF and can colonize solid substrates because their hypha can grow on particle surfaces and enter the inter-particle gaps (Patidar et al. 2018).
The CDH production by filamentous fungi under Submerged Fermentation has been studied extensively. However, there are very few reports on CDH production using SSF. In the present study, the organism was isolated and grown in SSF conditions to produce CDH by utilizing agro-industrial waste as a solid substrate. One factor at a time (OFAT) and Box-Behnken Design (BBD) of Response Surface Methodology (RSM) were used to statistically optimize the production.
MATERIAL AND METHODS
Isolation of fungi : Zip lock bags were used to collect samples from a variety of sources in Indore, India. The sources include air, damaged wood, soil, and municipal garbage. These samples were serially diluted, inoculated, and incubated on PDA media for 3 days at 28 °C. All isolates were preserved on PDA media for further screening.
Screening of potential CDH producer: The primary screening was carried out by growing isolates on Carboxymethyl cellulose (CMC) agar media containing (per liter) 10g CMC, 0.1g Yeast extract, 0.25g Peptone, 1.4g (NH4)2SO4, 2g KH2PO4, 0.3g MgSO4.7H2O, 0.3g Urea, 0.3g CaCl2, 3.34mg ZnSO4.7H2O, 5mg FeSO4.7H2O, 1.56mg MnSO4.7H2O, 2mg CoCl2 and 15g Agar, pH 5.0. It was inoculated at 28oC for 3 days (Shams et al. 2004). After incubation, 0.1% Congo red was flooded on agar media plates for 10 minutes and washed with 1 N NaCl. The fungi which formed a clear zone by flooding Congo red solution were selected and used for secondary screening (Amrinola et al. 2012).
The isolates selected from primary screening were grown in liquid media containing (per liter) 30g Microcrystalline cellulose, 30g Yeast extract, 1.0g MgSO4.7H2O and 0.3 ml trace element solution. Trace element solution contains (per liter), 0.3g MnCl2.4H2O, 3g H3BO3, 0.1g CuSO4.5H2O, 2g CoCl2.6H2O, 0.2g NiCl2.6H2O, 1g ZnSO4.7H2O and 4ml conc. H2SO4 (Ludwig et al. 2003; Fischer et al. 2014). The pH of the media was adjusted to 5. The cellulose-containing medium was prepared and divided into 100 ml aliquots in 250 ml Erlenmeyer flask and autoclaved at 121oC for 15 min. Each flask was inoculated with two agar plugs (6 mm diameter) taken from 5-day-old agar culture and was incubated at 150 rpm agitation condition at 30oC for 1 to 14 days (Baminger et al. 2001). Fungal culture supernatant was separated by centrifugation at 8000 rpm for 20 min at 4oC and used as a crude extract for enzyme assay.
Identification of fungal isolate: The fungal isolate W3 grown on PDA plates, was identified based on morphological and molecular characteristics. The growth characteristics and its microscopic observations were used to make a preliminary identification of the isolate. The molecular identification of the selected fungal isolate was done at the National Fungal Culture Collection of India (NFCCI), Pune, India. The Genomic DNA was isolated from W3 fungal isolate in pure form. The primers ITS4 & ITS5 were used for effective amplification of ITS-rDNA partial gene. The ABI-BigDye® Terminatorv3.1 Cycle Sequencing Kit was used to set up the sequencing PCR. The ABI 3100 automated DNA sequencer’s raw sequence was checked manually for consistency and compared to 18 r DNA sequences using the BLAST tool (Altschul et al. 1990). Closely related sequences were aligned using Clustal W software, and a phylogenetic tree based on the neighbor-joining (NJ) approach was created using the MEGA X program (Kumar et al. 2016).
Production of CDH by SSF : Preparation of inoculum: Schizophyllum commune was point inoculated on PDA media plate (90 mm diameter) and incubated for five days. Mycelia were scrapped from cultured plate after addition of 5ml sterile distilled water. Mycelia were collected and suspended in 10ml sterile distilled water and then vortexed with few glass beads of 0.1 mm diameter. The above fragmented mycelial culture was used as the inoculum for CDH production (Saha et al. 2008; Petrikkou et al. 2001).
Substrate selection : Agro-industrial residues mainly orange peel, wheat straw, banana peel, sugarcane bagasse, soybean straw, pineapple leaf, orange pulp, corn cob, groundnut shell, wood dust, coconut husk, and parthenium grass were screened for CDH production by SSF. These substrates were dried and crushed. SSF was performed using mineral solutions and distilled water to maintain a moisture content of 80% (v/w) while using 100 g of dry substrate in a 500 ml Erlenmeyer flask. The mineral solution contains (per liter) 26g KCl, 26g MgSO4, 76 g KH2PO4 and 0.3 ml trace element solution (as mentioned above in secondary screening) (Abdullah et al. 2016). All the flasks were autoclaved for 20 minutes at 121 °C. 1ml of inoculum was added to each flask and incubated at static condition at 30oC for 1 to14 days.
Enzyme extraction: The fermented medium (1g) was completely mixed with 100 mM sodium acetate buffer, pH 4.5 (1:15 w/v) and incubated at 30°C on a shaker at 120 rpm for 1 hr. The mixture was centrifuged at 5000xg for 15 min. at 4°C after being filtered using Whatman no. 1 filter paper. The enzyme assay was performed using the supernatant as a crude enzyme.
Enzyme assay and protein estimation: The reduction in absorbance of Dichloro Phenol Indo phenol (DCPIP) was used to determine CDH activity. The CDH activity assay was carried out by taking the reaction mixture containing 100 µl of 3mM DCPIP (in 10% ethanol), 100µl of 300mM lactose and 20µl of 80mM NaF in 100mM sodium acetate buffer (pH 4.5). The reaction was initiated by adding 100 µl of crude CDH. The decrease in absorbance at 520 nm was monitored for 10 minutes (Ludwig et al. 2003; Sulje et al. 2015). One unit of CDH activity has been defined as the amount of enzyme required for reduction of one µmol of DCPIP per min, under standard conditions. The amount of total protein was determined using the Folin Lowry method with bovine serum albumin (BSA) as standard (Waterborge and Metthews, 1994). All the experiments were performed in triplicate.
Optimization of CDH production using OFAT approach: OFAT method was used for the optimization of parameters such as initial moisture content (%), inoculum size (ml), temperature (oC), particle size (mm), and fermentation time (days). These parameters were the most effective variables representing a significant influence on CDH activity (Latifian et al. 2007; Darabzadeh et al. 2019).
Moisture content:The crushed groundnut shell was taken in Erlenmeyer flasks and moisture content was set in the range 30% to 90%, with interval of 10 % using the mineral solution and distilled water
(1:1). All the flasks were inoculated with 1 ml inoculum and incubated at 30oC. The crude enzyme was extracted after 8 days and enzyme assay was performed.
Inoculum size: The crushed groundnut shell was taken in Erlenmeyer flasks and inoculum size was set in the range 0.5 ml to 3 ml, with an interval of 0.5 ml. All flasks had 50% moisture content and were incubated at 30oC. The crude enzyme was extracted after 8 days and enzyme assay was performed.
Incubation temperature :The groundnut shell was taken in Erlenmeyer flasks, inoculated with 2 ml inoculum and 50% moisture content, and incubated at different incubation temperatures ranging from 20oC to 40oC with intervals of 5oC. The crude enzyme was extracted after 8 days and enzyme assay was performed in each sample.
Particle size :The groundnut shell of different particle size (0.5, 0.5-1 mm,1-1.5 mm and >2.0 mm) were taken in Erlenmeyer flasks. At this step, moisture content was 50%, inoculum size 2ml, and incubation temperature 30oC. The crude enzyme was extracted after 8 days and enzyme assay was performed.
Fermentation time : The prepared groundnut shell substrate was taken in Erlenmeyer flask and enzyme was extracted at different fermentation time ranging from 5 to 14 days. In this case, moisture content was 50%, inoculum 2ml, incubation temperature 30oC and particle size 0.5mm to1mm. The crude enzyme was extracted every 24 hours and enzyme assay was performed.
Statistical Analysis: After carrying out all the experiments in triplicate, the mean values for each experiment were calculated. Analysis of variance (ANOVA) was used to examine the data, and a significant result was defined as a P value of less than 0.05 (Nghi et al. 2021).
Statistical optimization of CDH production using RSM: OFAT method was used for the optimization of parameters such as moisture content (%), inoculum size (ml), incubation temperature (oC), particle size (mm), and fermentation time (days). Out of these, four variables namely moisture content, incubation temperature, particle size, and fermentation time were identified to be critical parameters in fermentation method. Response surface approach with BBD was used to examine the relationship between these important parameters and their optimum level (Box and Behnken, 1960).
Design-Expert 11.0 software was used to set up the experimental range, and each independent variable was examined at three different levels (+1,0, -1) using BBD. (Table 1). The following equation was used for the number of experiments (N) required for the creation of BBD:
N= 2k (k-1) + CO
Table 1. Independent variables used in BBD experimental design
| Variable | Name of parameters | Unit | Range and Levels | ||
| -1 | 0 | +1 | |||
| A | Fermentation time | Days | 5 | 8 | 11 |
| B | Moisture content | % (v/w) | 30 | 50 | 70 |
| C | Particle size of Groundnut shell | Mm | 0.5 | 1 | 1.5 |
| D | Temperature | oC | 25 | 30 | 35 |
Where, k denotes the number of variables and CO denotes the number of central points. By conducting 29 runs with five replicates at the centre point of experiment, this equation was used to construct a mathematical connection between four variables that are used in the production of CDH.
The following quadratic polynomial model equation was fitted using the response data:
Y = β0 + Σ βi Xi + Σ βii Xi2 + Σ βij Xij
Where, Y stands for predicted response; β0 for coefficient of fitted response; βi for linear coefficient; Xi for independent variables; βii for quadratic coefficient; Xij are variables interacting with each other and βij for interaction coefficient.
Table 2 denotes BBD’s experimental design in coded levels for the four variables. 2ml inoculum was added for all the experimental runs. The flasks were analysed for CDH production at regular time intervals, 5th, 8th and11th day, as designed by BBD.
Table 2. Process parameters for the production of CDH optimized using the BBD experimental design
| Standard Order | Run Order | Fermentation time | Moisture content | Particle size | Temperature | Actual value | Predicted value | ||||
| Coded | Decoded | Coded | Decoded | Coded | Decoded | Coded | Decoded | ||||
| 26 | 1 | 0 | 8 | 0 | 50 | 0 | 1 | 0 | 30 | 216.31 | 192.75 |
| 5 | 2 | 0 | 8 | 0 | 50 | -1 | 0.5 | -1 | 25 | 133.07 | 125.30 |
| 11 | 3 | -1 | 5 | 0 | 50 | 0 | 1 | +1 | 35 | 48.90 | 43.25 |
| 18 | 4 | +1 | 11 | 0 | 50 | -1 | 0.5 | 0 | 30 | 51.65 | 80.53 |
| 2 | 5 | +1 | 11 | -1 | 30 | 0 | 1 | 0 | 30 | 22.61 | 18.73 |
| 28 | 6 | 0 | 8 | 0 | 50 | 0 | 1 | 0 | 30 | 212.61 | 192.75 |
| 7 | 7 | 0 | 8 | 0 | 50 | -1 | 0.5 | +1 | 35 | 106.40 | 88.24 |
| 27 | 8 | 0 | 8 | 0 | 50 | 0 | 1 | 0 | 30 | 217.14 | 192.75 |
| 9 | 9 | -1 | 5 | 0 | 50 | 0 | 1 | -1 | 25 | 49.83 | 47.43 |
| 13 | 10 | 0 | 8 | -1 | 30 | -1 | 0.5 | 0 | 30 | 98.62 | 88.75 |
| 12 | 11 | +1 | 11 | 0 | 50 | 0 | 1 | +1 | 35 | 44.18 | 35.53 |
| 4 | 12 | +1 | 11 | +1 | 70 | 0 | 1 | 0 | 30 | 51.59 | 51.57 |
| 29 | 13 | 0 | 8 | 0 | 50 | 0 | 1 | 0 | 30 | 217.88 | 192.75 |
| 8 | 14 | 0 | 8 | 0 | 50 | +1 | 1.5 | +1 | 35 | 80.29 | 88.87 |
| 17 | 15 | -1 | 5 | 0 | 50 | -1 | 0.5 | 0 | 30 | 50.75 | 71.92 |
| 10 | 16 | +1 | 11 | 0 | 50 | 0 | 1 | -1 | 25 | 51.59 | 46.20 |
| 16 | 17 | 0 | 8 | +1 | 70 | +1 | 1.5 | 0 | 30 | 92.24 | 91.06 |
| 20 | 18 | +1 | 11 | 0 | 50 | +1 | 1.5 | 0 | 30 | 49.37 | 38.44 |
| 1 | 19 | -1 | 5 | -1 | 30 | 0 | 1 | 0 | 30 | 23.90 | 24.73 |
| 3 | 20 | -1 | 5 | +1 | 70 | 0 | 1 | 0 | 30 | 49.83 | 54.52 |
| 23 | 21 | 0 | 8 | -1 | 30 | 0 | 1 | +1 | 35 | 23.90 | 35.58 |
| 15 | 22 | 0 | 8 | -1 | 30 | +1 | 1.5 | 0 | 30 | 49.83 | 53.04 |
| 21 | 23 | 0 | 8 | -1 | 30 | 0 | 1 | -1 | 25 | 70.94 | 68.98 |
| 19 | 24 | -1 | 5 | 0 | 50 | +1 | 1.5 | 0 | 30 | 74.64 | 56.00 |
| 22 | 25 | 0 | 8 | +1 | 70 | 0 | 1 | -1 | 25 | 75.75 | 74.31 |
| 6 | 26 | 0 | 8 | 0 | 50 | +1 | 1.5 | -1 | 25 | 47.70 | 66.66 |
| 25 | 27 | 0 | 8 | 0 | 50 | 0 | 1 | 0 | 30 | 99.83 | 192.75 |
| 24 | 28 | 0 | 8 | +1 | 70 | 0 | 1 | +1 | 35 | 80.66 | 92.86 |
| 14 | 29 | 0 | 8 | +1 | 70 | -1 | 0.5 | 0 | 30 | 127.61 | 113.36 |
Validation of the experimental model: Under optimal conditions, the validation of the experimental model for CDH production was carried out in triplicates and the obtained findings were compared with the response predicted by the model.
RESULTS AND DISCUSSION
Isolation and Screening of potential CDH producer: A total of 108 isolates were obtained from various sources such as soil, air, water, degraded wood, dung, compost, and infected plant parts. All isolates were primarily screened on CMC-agar plate and diameter of clear zone in each case was determined. Out of all isolates, 12 isolates hydrolyzed the CMC-agar plates and showed the zone of clearance in a range from 3.8mm to 20mm (Table 3).
Table 3. Fungal isolates with zone of clearance of different diameter
| Isolate name | Zone diameter (mm) | Isolate name | Zone diameter (mm) |
| E1 | 20.1±1.0 | S10 | 9±2.2 |
| G4 | 19.83±3.0 | C2 | 8.1 ±1.5 |
| B2 | 16.1 ±1.0 | W6 | 7.1 ±1.5 |
| C8 | 15.5±2.2 | N1 | 6.8 ±3.0 |
| W7 | 11.5 ±1.7 | W3 | 5.1 ±0.5 |
| C6 | 10.3±1.0 | A2 | 3.8±0.5 |
The fungal isolates that produced a clear zone were selected for secondary screening. The secondary screening was used to confirm the CDH production in cellulose-containing liquid media under shaking conditions. W3 isolate showed maximum CDH activity. CDH activity was measured by a DCPIP-based assay. This fungal isolate was selected for further studies. Previous studies have reported that CDH is secreted by Phanerochaete chrysosporium (Bao et al. 1993), Monilia (Dekker et al. 1980), Sclerotium rolfsii (Sadana et al. 1988) and Schizophyllum commune (Fang et al. 1998). The zone diameter does not accurately corresponds to CDH production because the media contains CMC, which is not the specific substrate for CDH production (Amrinola et al. 2012) and the zone may be because of some other enzyme secretions. Similar results have been reported in Cladosporium isolates, where agar plate screening and liquid media cultivation were not parallel for CDH activity (Shams et al. 2004).
Table 4. ANOVA table showing the experimental results using the Box-Behnken design.
| Source | Sum of squares | Df | Mean square | F-value | p-value | |
| Model | 85068.24 | 14 | 6076.30 | 6.01 | 0.0009 | Significant |
| A-Fermentation time | 60.12 | 1 | 60.12 | 0.0594 | 0.8109 | |
| B-Moisture content | 2941.57 | 1 | 2941.57 | 2.91 | 0.1103 | |
| C-Particle size | 2523.87 | 1 | 2523.87 | 2.49 | 0.1366 | |
| D-Temperature | 165.39 | 1 | 165.39 | 0.1635 | 0.6921 | |
| AB | 2.33 | 1 | 2.33 | 0.0023 | 0.9624 | |
| AC | 171.22 | 1 | 171.22 | 0.1692 | 0.6870 | |
| AD | 10.50 | 1 | 10.50 | 0.0104 | 0.9203 | |
| BC | 45.02 | 1 | 45.02 | 0.0445 | 0.8360 | |
| BD | 674.70 | 1 | 674.70 | 0.6668 | 0.4278 | |
| CD | 877.94 | 1 | 877.94 | 0.8677 | 0.3674 | |
| A2 | 52656.29 | 1 | 52656.29 | 52.04 | <0.0001 | |
| B2 | 27632.78 | 1 | 27632.78 | 27.31 | 0.0001 | |
| C2 | 10868.09 | 1 | 10868.09 | 10.74 | 0.0055 | |
| D2 | 23004.58 | 1 | 23004.58 | 22.74 | 0.0003 | |
| Residual | 14165.06 | 14 | 1011.79 | |||
| Lack of Fit | 3355.06 | 10 | 335.51 | 0.1241 | 0.9964 | not significant |
| Pure Error | 10810.01 | 4 | 2702.50 | |||
| Cor Total | 99233.30 | 28 |
Identification of fungal isolate: Depending on morphological and molecular characteristics, the isolated W3 fungal strain was identified. Morphological identification of W3 fungal strain has shown that the isolate is filamentous and makes white cottony mycelia on PDA plate (Figure 2). The BLAST results of the ITS sequence (Figure 3) obtained from NCBI indicate the relationship of Schizophyllum commune BCC26414 with other isolates. There are many reports in the literature which are related to the identification of fungi based on ITS sequence (Raja et al. 2017). The tested fungal strain showed 99.83 % sequence similarity with Schizophyllum commune BCC26414, whose accession number is FJ372690. Previously reported Schizophyllum commune AS 5.391 also has the ability to produce CDH (Fang et al. 1998).
Figure 1: Congo red is used as an indicator on an agar plate with CMC as the substrate. The zone of clearance Surrounding the isolate revealed CMC degradation (a) control plate (b) W3 isolate

Figure 2: (a) W3 fungal isolate growth on potato dextrose agar. Microscopic view of Schizophyllum commune BCC26414 stained with lactophenol cotton blue at (b) 40 X (c)100 X in oil immersion in bright field microscopy.
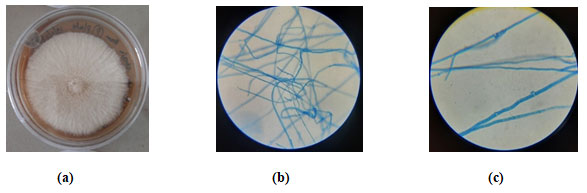
Figure 3: A phylogenetic tree created using the neighbor-joining technique that shows the connections between isolated strains. Accession numbers from the NCBI database of each isolate are given in the tree.

Figure 4: Substrate Selection for CDH production by Schizophyllum commune The data are
represented as the mean ±standard deviation (n=3), p<0.05.
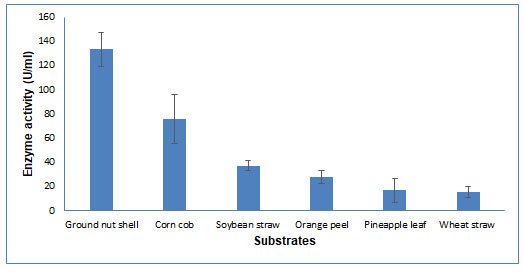
CDH production by Solid State Fermentation: Substrate selection: Several agro-waste materials were screened for CDH production using Schizophyllum commune. Out of these, six substrates (wheat straw, corn cob, groundnut shell, orange peel, soybean straw, and pineapple leaf) were utilized by the fungi in SSF for CDH production. Maximum CDH activity (133.3 U/ml) was obtained in groundnut shells and the least activity in wheat straw (15.25 U/ml) (Figure 4). According to Gupta et al. (2014), a cellulosic substrate with greater crystallinity may increase CDH production. (Gupta et al. 2014). The cellulose of groundnut shell has a high degree of crystallinity index (Manrich et al. 2021) and wheat straw has a low crystallinity index (Liu et al. 2005).
Figure 5: Influence of various moisture content on CDH production The data are
represented as the mean ±standard deviation (n=3), p<0.05.
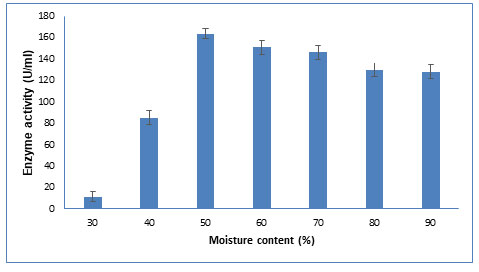
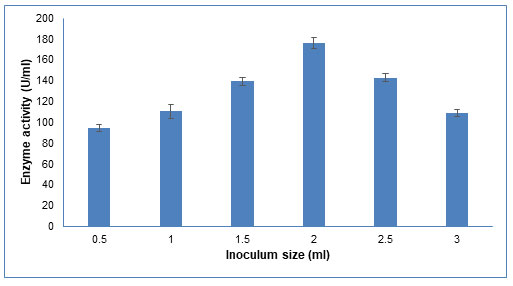
Figure 6: Influence of different inoculum sizes of Schizophyllum commune on CDH production
The data are represented as the mean ±standard deviation (n=3), p<0.05.
Tramitomycetes clypeatus grown in submerged fermentation on cellulose containing medium showed high CDH activity (55.88 U/mL) on eighth day at 30oC. Tramitomycetes clypeatus produced the highest yield of CDH activity when compared to other organisms that also used submerged fermentation, including Phanerochaete chrysosporium (0.8 U/ml), Schizophyllum commune (0.15 U/ml), and Sclerotium rolfsii (7.5 U/ml) (Saha et al. 2008). Very few reports on CDH production using SSF are available. CDH production has been reported earlier from Fusarium concolor using wheat straw (Li et al. 2008) and Coprinellus aureogranulatus using rice straw as a solid substrate in SSF (Nghi et al. 2021). This is the first report of CDH production by SSF using groundnut shells as a substrate.
Optimization of CDH production using OFAT approach: The one factor at a time (OFAT) method was used to optimise the growing conditions for the fungal isolate’s production of CDH.
Moisture content: The fermentation medium must have a proper level of moisture since it influences microbial development and biosynthesis. As shown in Figure 5, highest enzyme production was obtained at 50% moisture content. Similar results were observed for protease production using wheat bran and rice bran as a substrate in SSF with 50% moisture content (Chutmanop et al. 2008). As reported in other studies, low moisture content decreases nutrient solubility, water absorption, and substrate swelling, whereas excessive moisture content leads to the reduction of contact surface of fungus to the solid particles (Dutt and Kumar, 2014; Darabzadeh et al. 2019).
Inoculum size: An optimum inoculum size is required for enzyme production. As seen in Figure 6, the enzyme activity increased as the inoculum size increased from 0.5 ml to 2.0 ml. With further increase in inoculum size till 3.0 ml, the enzyme activity decreased. Similar results were reported by Gupta et al (2014), who found that increasing the inoculum size from 10% to 25% resulted in a decrease in CDH activity (Gupta et al. 2014). This could be because at low inoculum size, a longer lag phase is required, which results in lesser CDH activity. Upon increasing the inoculum size, rapid growth, and hence more enzyme production is favoured. However, a comparatively large inoculum size decreases enzyme production due to the quick depletion of nutrients (Gupta et al. 2014).
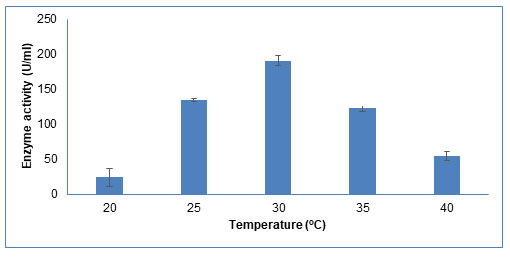
Temperature: The growth, physiology, and enzyme activity of microorganisms are strongly influenced by temperature. As shown in Figure 7, the enzyme activity increased as temperature increases from 20oC to 30oC. With further increase in temperature till 40oC, enzyme activity decreased. Highest enzyme activity at 30oC suggests its mesophilic nature. Saha et al (2008) also reported 30°C as optimum production temperature for CDH activity in Tramitomycetes clypeatus (Saha et al. 2008). This effect is due to the fact that at lower temperatures, substrate and product diffusion across the fungal cell is very less hence lowering the enzyme production. At elevated temperatures, the enzyme production declines due to its thermal denaturation (Dutt and Kumar, 2014; Lugani et al. 2015).
Figure 7: Influence of different incubation temperature on CDH production The data are
represented as the mean ±standard deviation (n=3), p<0.05.
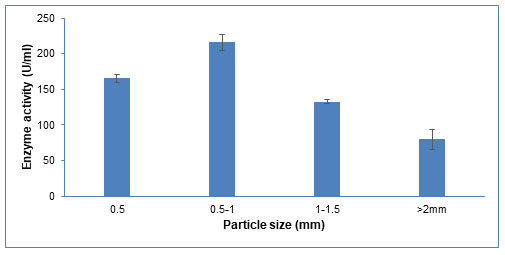
Particle size: Surface area is crucial for microbial adhesion and the mass transfer of different nutrients in SSF (Prakasham et al. 2006). As shown in Figure 8, the enzyme production increases upon increasing the particle size from 0.5mm to 1mm. A further increase in particle size till 2mm decreases the enzyme production. Similar results were observed by Matkawala et al (2019), where 1.4 mm particle size was considered optimum for alkaline protease production. An appropriate particle size provides an optimum surface area to facilitate microbial growth and product formation (Matkawala et al. 2019). Smaller substrate particles have more surface area, which promotes the development of microorganisms. However, too small substrate particles may cause substrate aggregation, which will slow the growth. Alternately, bigger particles size inhibits the growth of microorganisms because of their reduced surface area, lesser aeration, and mass transfer (Pandey et al. 2000).
Figure 8: Influence of different particle size of groundnut shell on CDH production.
The data are represented as the mean ±standard deviation (n=3), p<0.05.
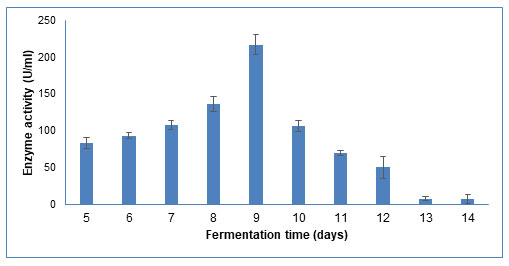
Fermentation time: The fermentation time is associated with microbial growth and enzyme production. As shown in Figure 9, the enzyme activity increased up to 9th day, after which it decreased as observed till 14th day. Similar results have been reported in Tramitomyces clypeatus, where maximum CDH production was found on 8th day of submerged fermentation, at 30oC under shaking conditions (Saha et al. 2008).
Figure 9: Influence of different incubation time on CDH production by Schizophyllum commune
The data are represented as the mean ±standard deviation (n=3), p<0.05.
Statistical Analysis : Statistical optimization of CDH production using RSM: By using the OFAT method, the impact of different process parameters on CDH production was investigated. Four components, including moisture content, incubation temperature, particle size, and fermentation time significantly affected the enzyme production. Response surface analysis and BBD were used to establish the optimal level of significant factors to enhance CDH production by Schizophyllum commune.
Using a set of 29 experiments, the effects of four independent variables were examined at three levels and five central points. The results are shown in Table 2. Using Design-Expert 11.0 software, analysis of variance (ANOVA) was performed to determine the statistical significance of the model equation. The results are shown in Table. 4. A second-order polynomial equation was used to fit the obtained response:
Y=2811.77 + 166.396 A + 12.7667 B + 138.796 C + 130.629 D + 0.0127083 AB – 4.36167 AC- 0.108 AD + 0.3355 BC + 0.129875 BD + 5.926 CD – 10.011 A2 – 0.163173 B2 – 163.731 C2 – 2.38211 D2
Where A represents fermentation time, B represents moisture content, C represents particle size and D represents incubation temperature. The effect of experimental variables on enzyme production was correlated using this equation. Multiple linear regressions were used to estimate the model coefficients, and those with (P<0.05) were determined to be significant. The experimental design revealed that the highest CDH activity was 217 U/ml at 50% moisture, 8th day of fermentation, 1 mm groundnut shell particle size, and 30oC temperature. These results revealed that the predicted and experimental values did not differ significantly, indicating that the model is appropriate for maximizing CDH production. The model F-value of 6.01 indicated that the model is significant (P< 0.0009).
The “Lack of Fit F-value” of 0.1241 indicates that the data fits the model and that the Lack of Fit is not statistically significant when compared to pure error, which is the desired characteristic. The determination coefficient was used to evaluate the model’s quality of fit (R2). In this investigation, the model’s R2 value was determined to be 0.857; the adjusted and predicted R2 values were computed as 0.714 and 0.635, respectively, with a difference of less than 0.2. This showed that this model can account for only 0.08% of the overall variations. As a result, the current R2 value shows that the trial runs were accurate and consistent, and the model is reliable for CDH production.
Response surface plot: Response surface plots demonstrate the interactions of variables (Figure 10). Each response surface plot in this instance depicts the impact of two independent variables while maintaining the levels of the other variables at zero.When the effect of fermentation time and moisture content were plotted against the enzyme activity, a link was seen; as a result, increasing moisture content with extended fermentation times encourages reaction up to the optimal level (Figure 10a). Increases in moisture content of the solid state medium up to 50% were shown to increase CDH production, which was followed by decrease in enzyme production. Similar to this, de Castro et al. (2015) employed 50% moisture to produce protease using Aspergillus niger (de Castro et al. 2015). The response plot revealed that extending the fermentation period until 8th day favored maximum CDH production by Schizophyllum commune. Fermentation time plays a significant role in commercial production of enzymes.
Figure 10: Response surface plots illustrating the interactions between (a) Fermentation time and moisture content (b) Fermentation time and particle size (c) Fermentation time and incubation temperature (d) Moisture content and particle size (e) Incubation temperature and moisture content (f) Incubation temperature and particle size.
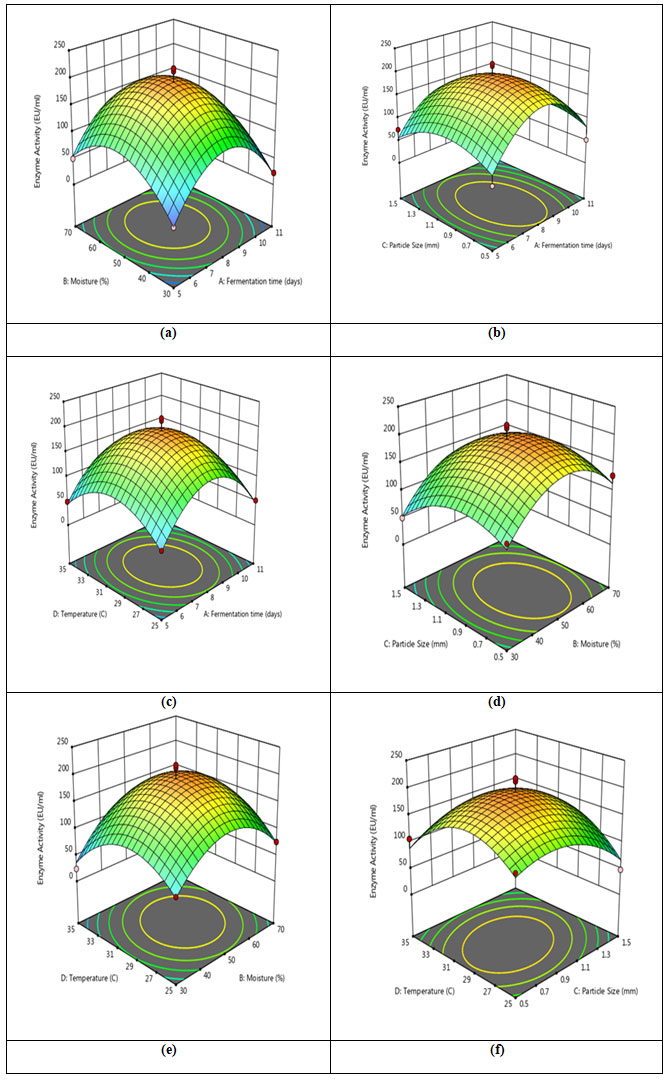
The optimal temperature was found to be 30oC for the production of CDH based on the response surface plots, which indicated that increasing the temperature enhanced enzyme production. (Figure 10c, 10e, 10f). Similar findings were observed when Aspergillus niger produced xylanolytic enzyme in submerged fermentation at 30 °C using RSM methods for optimization (Pellieri et al. 2022).
The response surface plots (Figure 10b, 10d, and 10f) represent the interaction between particle size of groundnut shell with fermentation time, moisture content and temperature, respectively. Here, increasing groundnut shell particle size initially enhanced CDH synthesis; however, an optimum level was predicted at 1 mm, and after that reduction was seen. Although the particle size of ground nut shells and temperature were found to have a slightly inclined curve (Figure 10f), the graph shows that there is only minor interaction between these variables and particle size. Similarly, Prakasham et al (2006) reported 1.0 to 1.4 mm optimum particle size of green husk for protease production (Prakasham et al. 2006).
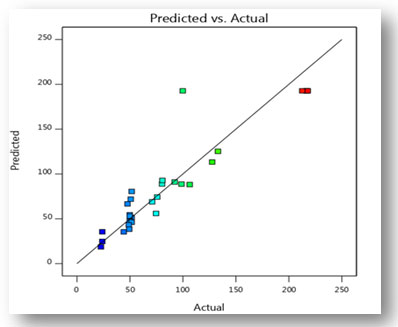
Validation of the experimental model: Fermentation was carried out under the predicted conditions to validate the experimental model. The optimum values of four variables under consideration are fermentation time 8th day, moisture content 50%, particle size 1mm and temperature 30oC. Under optimal conditions, CDH production was predicted to be 192.75 (U/ml), however, it was actually measured at 217.88 (U/ml). There were many similarities between the actual and predicted experimental results, hence the proposed model is accurate and highly successful. The optimization of different production variables by statistical method resulted in 1.63-fold enhancement in the CDH production, as compared to results (133.3 U/ml) obtained in unoptimized conditions.
Figure 11: Graph illustrating the relationship between predicted and actual values serves as
experimental validation of the CDH production model.
OFAT approach is difficult as it involves multiple parameters which need to be managed simultaneously, hence RSM is the preferred method for optimization (Lahiri et al. 2021). CDH isolated from Coprinellus aureogranulatus was used for the degradation of rice straw, where fungal hydrolases and metabolites were identified and further optimized using the RSM statistical approach (Nghi et al. 2021). Nawawi et al (2022) used Central Composite Design (CCD) model to enhance 1.34-fold xylanase and 5.96-fold pectinase production as compared to OFAT approach (Nawawi et al. 2022). Alves et al (2022) investigated optimum conditions for the multi-enzymatic recovery of cellulases produced by Aspergillus niger using sugarcane bagasse.
The ultrasound effects were evaluated using a Doehlert design while temperature, time, and pH were analyzed using the Box–Behnken design (Alves et al. 2022). Box-Behnken Design was adopted by Mishra (2016) to optimize the fermentation conditions using Brevibacillus brevis and a 1.5-fold increase in protease production was achived (Mishra, 2016). Gupta et al (2014) observed that both organic and inorganic nitrogen sources affected CDH production, the optimization of these nitrogen sources was carried out using BBD (Gupta et al. 2014). The present study is the first ever contribution to the use of groundnut shell as a solid substrate for the production of CDH using Schizophyllum commune and its statistical optimization.
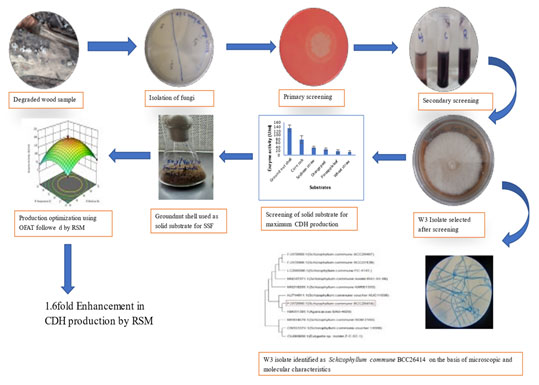
Graphical abstract
CONCLUSION
In the present study, Schizophyllum commune BCC26414 has been used for CDH production by solid-state fermentation (SSF). CDH production has been optimized and maximum CDH production was obtained when groundnut shell was used as a substrate at 30°C on the 9th day of incubation with initial moisture content 50%, using Schizophyllum commune BCC26414. This is the first report on CDH production and its statistical optimization by using groundnut shell as solid substrate. The groundnut shell is a waste from groundnut, abundantly cultivated in India. In 2020, groundnut production in Madhya Pradesh was 0.35 million tonnes. A potential strain like Schizophyllum commune BCC26414 can efficiently use this biowaste to produce the commercially significant CDH. Hence this study can be useful in the cost-effective production of CDH for various applications.
ACKNOWLEDGMENTS
The work is supported by CSIR-JRF fellowship program. The National Fungal Culture Collection of India (NFCCI), located in Pune, is acknowledged by the authors for its resources for fungal identification. The authors also acknowledge the facilities of the School of Biotechnology DAVV Indore, used in the present study.
Declaration: The authors state that they have no competing interests, either financial or otherwise.
REFERENCES
Abdullah, J. J., Greetham, D., Pensupa, N., et al. (2016). Optimizing cellulase production from municipal solid waste (MSW) using solid state fermentation (SSF). Journal of Fundamentals of Renewable Energy and Applications, 6(3), 1-10.
Altschul, S. F., Gish, W., Miller, W., et al. (1990). Basic local alignment search tool. Journal of molecular biology, 215(3), 403-410.
Alves, T. P., Triques, C. C., da Silva, E. A., et al. (2022). Multi‐enzymatic recovery of fungal cellulases (Aspergillus niger) through solid‐state fermentation of sugarcane bagasse. The Canadian Journal of Chemical Engineering, 100(8), 1930-1940.
Amrinola, W., Hasanah, N., Prasetya, B., et al. (2012). Isolation and screening of fungi producing cellobiose dehydrogenase:” enzymes for animal feed preparations based on enzymatic process”. International Journal of Pharma and Bio Sciences, 3(1).
Baminger, U., Subramaniam, S. S., Renganathan, V., et al. (2001). Purification and characterization of cellobiose dehydrogenase from the plant pathogen Sclerotium (Athelia) rolfsii. Applied and environmental microbiology, 67(4), 1766-1774.
Bao, W. J., Usha, S. N., and Renganathan, V. (1993). Purification and characterization of cellobiose dehydrogenase, a novel extracellular hemoflavoenzyme from the white-rot fungus Phanerochaete chrysosporium. Archives of biochemistry and biophysics, 300(2), 705-713.
Banerjee, S., and Roy, A. (2021). Molecular cloning, characterisation and expression of a gene encoding cellobiose dehydrogenase from Termitomyces clypeatus. Gene Reports, 23, 101063.
Box, G. E., and Behnken, D. W. (1960). Some new three level designs for the study of quantitative variables. Technometrics, 2(4), 455-475.
Cameron, M. D., and Aust, S. D. (2001). Cellobiose dehydrogenase–an extracellular fungal flavocytochrome. Enzyme and microbial technology, 28(2-3), 129-138.
Chutmanop, J., Chuichulcherm, S., Chisti, Y., et al. (2008). Protease production by Aspergillus oryzae in solid‐state fermentation using agroindustrial substrates. Journal of Chemical Technology & Biotechnology: International Research in Process, Environmental & Clean Technology, 83(7), 1012-1018.
Darabzadeh, N., Hamidi‐Esfahani, Z., and Hejazi, P. (2019). Optimization of cellulase production under solid‐state fermentation by a new mutant strain of Trichoderma reesei. Food science & nutrition, 7(2), 572-578.
de Castro, R. J. S., Ohara, A., Nishide, T. G., et al. (2015). A versatile system based on substrate formulation using agroindustrial wastes for protease production by Aspergillus niger under solid state fermentation. Biocatalysis and agricultural biotechnology, 4(4), 678-684.
Dekker, R. F. (1980). Induction and characterization of a cellobiose dehydrogenase produced by a species of Monilia. Microbiology, 120(2), 309-316.
Dutt, D., and Kumar, A. (2014). Optimization of cellulase production under solid-state fermentation by Aspergillus flavus (AT-2) and Aspergillus niger (AT-3) and its impact on stickies and ink particle size of sorted office paper. Cell Chem Technol, 48(3-4), 285-298.
Fang, J., Liu, W., and Gao, P. J. (1998). Cellobiose dehydrogenase fromSchizophyllum commune: Purification and study of some catalytic, inactivation, and cellulose-binding properties. Archives of Biochemistry and Biophysics, 353(1), 37-46.
Fischer, C., Krause, A., and Kleinschmidt, T. (2014). Optimization of production, purification and lyophilisation of cellobiose dehydrogenase by Sclerotium rolfsii. BMC biotechnology, 14(1), 1-12.
Gangwar, R., Rasool, S., and Mishra, S. (2021). Purified cellobiose dehydrogenase of Termitomyces sp. OE147 fuels cellulose degradation resulting in the release of reducing sugars. Preparative Biochemistry & Biotechnology, 51(5), 488-496.
Gupta, G., Gangwar, R., Gautam, A., et al. (2014). Production of cellobiose dehydrogenase from a newly isolated white rot fungus Termitomyces sp. OE147. Applied biochemistry and biotechnology, 173(8), 2099-2115.
Henriksson, G., Johansson, G., and Pettersson, G. (2000). A critical review of cellobiose dehydrogenases. Journal of biotechnology, 78(2), 93-113.
Kumar, S., Stecher, G., & Tamura, K. (2016). MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Molecular biology and evolution, 33(7), 1870-1874.
Lahiri, D., Nag, M., Mukherjee, D., et al. (2021). Recent trends in approaches for optimization of process parameters for the production of microbial cellulase from wastes. Environmental Sustainability, 4(2), 273-284.
Latifian, M., Hamidi-Esfahani, Z., and Barzegar, M. (2007). Evaluation of culture conditions for cellulase production by two Trichoderma reesei mutants under solid-state fermentation conditions. Bioresource technology, 98(18), 3634-3637.
Li, L., Li, X. Z., Tang, W. Z., Zhao, J., and Qu, Y. B. (2008). Screening of a fungus capable of powerful and selective delignification on wheat straw. Letters in Applied Microbiology, 47(5), 415-420.
Liu, R., Yu, H., and Huang, Y. (2005). Structure and morphology of cellulose in wheat straw. Cellulose, 12(1), 25-34.
Ludwig, R., and Haltrich, D. (2003). Optimisation of cellobiose dehydrogenase production by the fungus Sclerotium (Athelia) rolfsii. Applied microbiology and biotechnology, 61(1), 32-39.
Lugani, Y., Singla, R., and Sooch, B. S. (2015). Optimization of cellulase production from newly isolated Bacillus sp. Y3. Journal of bioprocessing & biotechniques, 5(11), 1.
Manrich, A., Martins, M. A., and Mattoso, L. H. C. (2021). Manufacture and performance of peanut skin cellulose nanocrystals. Scientia Agricola, 79.
Matkawala, F., Nighojkar, S., Kumar, A., and Nighojkar, A. (2019). Enhanced production of alkaline protease by Neocosmospora sp. N1 using custard apple seed powder as inducer and its application for stain removal and dehairing. Biocatalysis and Agricultural Biotechnology, 21, 101310.
Mishra VK (2016) Optimization of thermotolerant alkaline protease production from Brevibacillus brevis strain BT2 using surface response methodology. Biocatalysis and Agricultural Biotechnology. 7:87-94.
Mrudula, S., and Murugammal, R. (2011). Production of cellulase by Aspergillus niger under submerged and solid-state fermentation using coir waste as a substrate. Brazilian Journal of Microbiology, 42, 1119-1127.
Nawawi, M. H., Ismail, K. I., Sa’ad, N., et al. (2022). Optimisation of Xylanase–Pectinase Cocktail Production with Bacillus amyloliquefaciens ADI2 Using a Low-Cost Substrate via Statistical Strategy. Fermentation, 8(3), 119.
Nghi, D. H., Kellner, H., Büttner, E., et al. (2021). Cellobiose dehydrogenase from the agaricomycete Coprinellus aureogranulatus and its application for the synergistic conversion of rice straw. Applied Biological Chemistry, 64(1), 1-11.
Nyanhongo, G. S., Thallinger, B. and Guebitz, G. M. (2017). Cellobiose dehydrogenase-based biomedical applications. Process Biochemistry, 59, 37-45.
Pandey, A., Soccol, C. R., Nigam, P., et al. (2000). Biotechnological potential of coffee pulp and coffee husk for bioprocesses. Biochemical Engineering Journal, 6(2), 153-162.
Patidar, M. K., Nighojkar, S., Kumar, A., and Nighojkar, A. (2018). Pectinolytic enzymes-solid state fermentation, assay methods and applications in fruit juice industries: a review. 3 Biotech, 8(4), 1-24.
Pellieri, C. M., Taddia, A., Loureiro, D. B., et al. (2022). Spartina argentinensis valorization and process optimization for enhanced production of hydrolytic enzymes by filamentous fungus. Environmental Technology & Innovation, 26, 102298.
Petrikkou, E., Rodrı́guez-Tudela, J. L., Cuenca-Estrella, M., et al. (2001). Inoculum standardization for antifungal susceptibility testing of filamentous fungi pathogenic for humans. Journal of clinical microbiology, 39(4), 1345-1347.
Prakasham, R. S., Rao, C. S., and Sarma, P. N. (2006). Green gram husk—an inexpensive substrate for alkaline protease production by Bacillus sp. in solid-state fermentation. Bioresource technology, 97(13), 1449-1454.
Rai, R., Basotra, N., Kaur, B., Di Falco, M., Tsang, A., and Chadha, B. S. (2020). Exoproteome profile reveals thermophilic fungus Crassicarpon thermophilum (strain 6GKB; syn. Corynascus thermophilus) as a rich source of cellobiose dehydrogenase for enhanced saccharification of bagasse. Biomass and bioenergy, 132, 105438.
Raja, H. A., Miller, A. N., Pearce, C. J., and Oberlies, N. H. (2017). Fungal identification using molecular tools: a primer for the natural products research community. Journal of natural products, 80(3), 756-770.
Sadana, J. C., and Patil, R. V. (1988). Cellobiose dehydrogenase from Sclerotium rolfsii. In Methods in Enzymology (Vol. 160, pp. 448-454). Academic Press.
Saha, T., Ghosh, D., Mukherjee, S., Bose, S., and Mukherjee, M. (2008). Cellobiose dehydrogenase production by the mycelial culture of the mushroom Termitomyces clypeatus. Process Biochemistry, 43(6), 634-641.
Shams Ghahfarokhi, M., Fazli, A., Lotfi, A., and Razzaghi Abyaneh, M. (2004). Cellobiose dehydrogenase production by the genus Cladosporium. Iranian Biomedical Journal, 8(2), 107-111.
Sulej, J., Janusz, G., Osińska-Jaroszuk, M., et al. (2015). Characterization of cellobiose dehydrogenase from a biotechnologically important Cerrena unicolor strain. Applied biochemistry and biotechnology, 176(6), 1638-1658.
Waterborg, J. H., and Matthews, H. R. (1994). The Lowry method for protein quantitation. Basic protein and peptide protocols. Humana, 1, 1-4.


