1Department of Biological Science, King Abdulaziz University, Jeddah, Saudi Arabia,
2Department of Microbiology, King Abdulaziz University, Rabigh, Saudi Arabia.
3Botany and Microbiology Department, Faculty of Science, Kafrelsheikh University, Egypt
Corresponding author email: magdammali@hotmail.com
Article Publishing History
Received: 11/12/2020
Accepted After Revision: 24/03/2021
Under harsh environmental stresses, some plants can survive due to their association with microorganisms. These microorganisms residing within the specific host plant, form unique group of endophytes that can extend diverse sorts of positive impacts on plant. Endophytes related to their functions to host plant and factors affecting their association are poorly studied. Thus, this study concentrated onisolation, identification, and characterization of endophytic bacteria from Heliotropium pterocarpum growing at Hot spring, Gomyqah, Saudi Arabia, for their biological impacts as plant growth promoting (PGP) traits. Seven endophytic bacteria were isolated from root, stem, and leave of H. pterocarpum plant. These bacterial isolates were molecular identified based on 16S rDNA, as Serratia sp., Exigobacterium indicum, Kocuria sediminis, Variovorax paradoxus, Staphylococcus epidermis, Siphingobium yanoikuyae and Serratia rubidea.
In addition, 42% of these bacterial isolates have efficacy of phosphate solubilizing with clear zones ranging from 5.2 to 6.6 mm, and siderophore producing ranging from 14 to 16.3 mm.Moreover,most of thesebacterial isolates have ability to producedifferent enzyme activities. Furthermore, all the selected bacterial isolates were able to produceIndole acetic acid (IAA) and Gibberellic acid (GA3) in broth media, ranging from 0.002 to 0.056 mg/ml, and from 0.006 to 0.144 mg/ml, respectively. Considering all these activities of bacterial isolates,endophytes could be exploited as effective resource for promoting plant growth and nutrient uptake without chemical effecton the environment. Thus, endophytic bacteria could be used as biological productin agriculture fields.
Heliotropium pterocarpum; Endophytic, Bacteria; 16S Rdna, Extracellular Enzymes
Ashkan M, Aly M, Aldhebiani A. Screening and Characterization of Endophytic Bacteria from Heliotropium pterocarpum Growing at hot Spring for their Biological Impacts. Biosc.Biotech.Res.Comm. 2021;14(1).
Ashkan M, Aly M, Aldhebiani A. Screening and Characterization of Endophytic Bacteria from Heliotropium pterocarpum Growing at hot Spring for their Biological Impacts. Biosc.Biotech.Res.Comm. 2021;14(1). Available from: <a href=”https://bit.ly/3cXEZ4H“>https://bit.ly/3cXEZ4H</a>
Copyright © Ashkan et al., This is an open access article distributed under the terms of the Creative Commons Attribution License (CC-BY) https://creativecommns.org/licenses/by/4.0/, which permits unrestricted use distribution and reproduction in any medium, provide the original author and source are credited.
INTRODUCTION
Under extreme environmental stresses, plants can survive by developing symbiotic association with any components of their ecosystem to survive and development in their natural environments. Microorganisms are one of the most vital components that create useful associations with plants (Santoyo et al., 2016). Symbiotic bacteria or “endophytes” can associate internally with a wide-range of plants species (Guo et al., 2008). Endophytes defined as a group of microorganisms that colonizing internally different parts of host plant, without causing symptomatic impacts to the plant (Wilson, 1995).
Endophytes distribution within plants depends on their ability to colonize and obtain plant resources. Endophytic metabolic pathways played major roles in endophytic diversity (Conrath et al., 2006; Singh et al., 2009) which were contained mainly four phyla: Proteobacteria, Actinobacteria, Firmicutes, and Bacteroidetes, with dominant genera of Pseudomonas, Bacillus, Burkholderia, Stenotrophomonas, Micrococcus, Pantoea and Microbacterium (Goryluk-Salmonowicz et al., 2018).
Researchers are supposed that presence of these endophytes improveplant ecology by increasing the capacity of host plants to grow under environmental and biological stresses (Miliute et al., 2015). It is believed that endophytes live in close association with plants and can extend different kinds of positive effects on plant growth and health. These include increased nutrient availability through the fixation of nitrogen, solubilization of phosphate, and production of siderophores.
Furthermore, endophytes could improve plant through production of plant growth regulators phytohormones, such as indole-3-acetic acid (IAA), gibberellic acid (GA3), and cytokinins in addition to their ability to tolerate stress, drought, salinity, and metal toxicity (Nogema et al., 2013). Endophytes also could provide resistance to diseases and potential pathogens through production of secondary metabolites and extracellular enzymes (Nair and Padmavathy, 2014). Moreover, the mechanisms of endophytes to deal with natural ecology may give superior survival preferences to host plants.
Bacteria of endophytic plants were first studied in temperate regions, but recently the studies were extended to plants in tropical regions as well. However, a little knowledge is available on diversity, and interaction of endophytes communities in plants struggling for existence in extreme environment. Thus, focusing on isolating and screening of endophytes with diverse properties from plants at Hot spring are necessary and urgent. These endophytes could have many roles to manipulate plant growth (Patten and Glick 2002, Fadiji and Babalola, 2020).
Using these interesting of endophytes with many roles are useful especially in poor soil and desert regions. Heliotropium pterocarpum is widely spread at Hot spring, Gomyqah (Saudi Arabia). However; the biological properties of this plant related to their endophytic biology have not yet been well studied. Hence, this study was focused on isolation, identification, and characterizing of endophytic bacteria from the previous plant and screening these endophytes for production of plant growth regulators and other promoting materials to understand their functional roles.
MATERIAL AND METHODS
Plant collection from the study area: This study was carried out at Hot spring area (N 40˚28’11.4E), located 17 Km North-East of Gomyqah city, and 25 km East of Al-Leeth city, Saudi Arabia. Sterile technique was used during the collection of plant specimens. The plantwas collected, labeled, and immediately transferred in sterile polyethylene bags to the Microbiological laboratory at faculty of Science, andplaced on a dry cool place to avoid moisture accumulation or excessive drying (stored at 4°C). The plants were collected from 2 meters distance of a stream bank at Hot spring area, during February 2019 at 32°C for the isolation of endophytes.
Identification of collected plants: Plant sample was identified by Dr. Amal Aldhebiani (Botany/Biology Department, King Abdulaziz University, Jeddah, SA).The plant sample was characterized, identified and checked with some books and online checklist of the Flora of KSA (Alfarhan and Thomas,1994;Chaudhary, 2001), as well as with Thomas (2011). Then, nomenclature and family of the plant was identified and checked by follow Catalogue of Life website.
Surface sterilization and isolation of bacterial endophytes: To remove dust and debris, the plant parts were firstly washed with water, then surface sterilized by following sterilization- culturing dependent method (Hallmann et al. 1997; Zinniel et al. 2003), using 70% ethanol and aqueous solution of 5% sodium hypochlorite, rinsed with sterile distilled water and dried using sterile filter paper. Sterilized surgical blade was used to cut the plant into small pieces (1-3 mm long) and each parts of plant were put on Nutrient Agar plate (Hi Media, Mumbai, India).
To confirm the disinfection process, aliquots of the steriledistilled water that used in the final plant rinse was plated onto the previous medium. All plates were incubated at 28˚C for 1-2 weeks to allow growth of endophytes and examined for bacterial endophytic growth from the used plant segments. Bacteria growing out of the plant segments were isolated, purified, and identified based on phenotypic and genotypic characteristics.
Molecular identification of endophytes bacterial isolates: The bacterial isolates were identified by extracting the genomic 16S rDNA (Govindarajan et al., 2007) from the bacterial colonies, using a commercial kit for bacterial DNA extraction (MQ Bacterial DNA Isolation Kit, MOLEQULE-ON Company, Auckland, New Zealand).
Then, 16SrDNA was amplified in using the genomic DNA as template and bacterial universal primers, 27 F (5′GAGTTTGATCCTGGCTCA-3′), and reverse primer 1492R (5’-GGTTACCTTGTTACGACTT-3’). The PCR product was visualized, sequenced at Macrogen Online Sequencing Company, Korea and then, checked by BLAST analysis in the NCBI database for microbial identification. The 16S rDNA sequence of the strains was used to search the GenBank database and determine phylogenetic relative strains.
Screening of endophytic bacterial isolates for biological impacts: Solubilization of phosphate: The bacterial isolates were screened for phosphate solubilization using Pikovskaya’s medium (pH 7.0) which composed of (g/l): glucose 10; tri-calcium phosphate 5; ammonium sulphate 0.5; sodium chloride 0.2; magnesium sulphate heptahydrate 0.1; potassium chloride 0.2; ferrous sulfate heptahydrate 0.002; yeast extract 0.5; manganese
(II) sulfate dehydrate 0.002; bromo phenol 0.025g, Bacto agar (Difco) 15. The medium was inoculated by spotting 10 μl of overnight shaken bacterial broth cultures on the surface of Pikovskya agar and the plates were incubated at 28°C for 3-5 days. Formation of a clear halo zone around the colony was due to the utilization of tricalcium phosphate present in the medium (Lavakush and Verma, 2012).
Production of siderophores: Siderophores produced by the endophytic isolates were determined using qualitative assay as described by Schwyn and Neilands (1985) using chrome azurol S (CAS), and hexadecyltrimethylammonium bromide (HDTMA) as indicators. The CAS/ HDTMA react with ferric iron to produce a blue color. Removing a siderophore (iron chelator) from the dye complex changes the color from blue to orange. On each plate of CAS medium, 10 μl of 48 hours old cultures of endophytic bacterial filtrate were spotted and all plates were incubated at 28ºC for 2 days. A color change of the CAS medium around the colony from blue to yellow was considered positive result (Louden et al., 2011).
Exoenzyme activity: The extracellular hydrolytic enzyme activity was detected by growing the bacterial isolates on different indicator media, including amylase activity medium (Glucose Yeast Extract Peptone Agar (GYP) medium containing 2.0% (w/v) starch and 1.5% agar (w/v)(Claus 1988), lipas eactivity medium (NB medium containing 1.0% Tween 80 (v/v) and 1.5% agar (w/v) (Rajanet.al, 2011), protease activity medium(NB medium
containing 1.0% (w/v) skim milk and1.5% agar (w/v) (Tennalli et al, 2012), pectinase activity medium (NB medium containing 0.5% poly galaturonate (v/v) and 1.5% agar (w/v) (Cotty et al. 1990), cellulase activity medium (carboxy methyl cellulose (CMC) medium containing 0.5% (w/v) carboxyl methyl cellulose and 1.5% agar (w/v) (Zaghloul et al. 2016). All the isolates were spotted inoculated on respective enzymes screening media and incubated at 28ºCfor 48-72 hours. Clearing zones in the medium indicated positive enzyme activity.
Production of Indole Acetic Acid and Gibberellic Acid: Estimation of IAA was recorded using a colorimetric spectrophotometric method (Patten and Glick, 2002). Each endophytic bacterial isolate was grown in250 ml flasks Erlenmeyer flask containing 50 ml of nutrient broth containing 0.2 % of L-tryptophan (v/v). After incubation in darkness for 7days at 30℃and 120 rpm,the culture filtrate was centrifuged at 10,000 rpm for 15 min., and then 2 ml of each culture supernatant was mixed with 2 drops of concentrated orthophosphoric acid, followed by 4 ml of Salkowski reagent.
The mixture was incubated in darkness at room temperature (25℃) for 25 min, and the presence of pink color indicated IAA production. The absorbance was read at 530 nm using a spectrophotometer (SpectroSC™ Spectrophotometer, LaboMed.inc). A standard curve of known concentrations of IAA was prepared to determine the quantities of IAA in each filtrate. Similarly, the amount of GA3 produced by the endophyte’s isolates was estimated by the method of Holbrook et al., (1961).
Two ml of zinc acetate solution was added to 50 ml of the bacterial culture filtrate and after two minutes of incubation, two ml of potassium ferrocyanide solution was added, the reaction volume was centrifuged at 8000 rpm for 10 minutes. Five ml of supernatant was added to five ml of 30 % HCl and the mixture was incubated at 28ºC for 75 min. Five ml of the supernatant with five ml of 5% HCl was used as blank. The absorbance of the sample and blank were measured at 254 nm. A standard curve was prepared by using gibberellic acid to calculate the GA3 quantities in each bacterial filtrate (Holbrook et al., 1961).
RESULTS AND DISCUSSION
Hot spring at Gomyqah, Al-Leeth city, Kingdom of Saudi Arabia (Figure 1- A) was visited and a plant from the normal flora was two meters distance far away from the stream bank of Hot spring (Figure 1-B). This collected plant was identified as Heliotropium pterocarpum (A. DC.) Hochst. & Steud. ex Bunge (Figure 1-C), which belong to Borage family (Boraginaceae) (Table 1). Plant sample was mounted on sheet bearing a label and saved at King Abdulaziz University herbarium.
Figure 1: A: Google map of the Hot spring area at Gomyqah village at Al-Leeth city, B: The study area, C: The collected H. pterocarpum at natural habitat

Table 1. The scientific name and classification of Heliotropium pterocarpum plant according to catalog of life website.
| Accepted scientific name | Heliotropium pterocarpum (A. DC.) Hochst. & Steud. ex Bunge (accepted name). |
| Basionym | Heliophytum pterocarpum A. DC. in DC. (1845, p. 552). |
| Taxonomic synonym | Bourjotia pterocarpa (DC.) Pomel, Heliophytum pterocarpum DC. & A. DC, and Heliotropium kassasii Täckh. & Boulos. |
A total of 7 endophytic bacterial isolates were obtained from H. pterocarpum. The bacterial isolates namely P1M4, P1M6, P1M7, P1M8, P1M9 were isolated from the leaves, and P1SM10 and P1SM11 were isolated from the stem segments of H. pterocarpum. These isolateswere morphologically characterized by Gram staining test, 57% were Gram negative, rods shape, while 43% were Gram positive, bacilli and cocci in shape. The DNA sequences were analyzed by BLAST analysis for alignment,the results were compared with NCBI database and phylogenic tree were obtained.
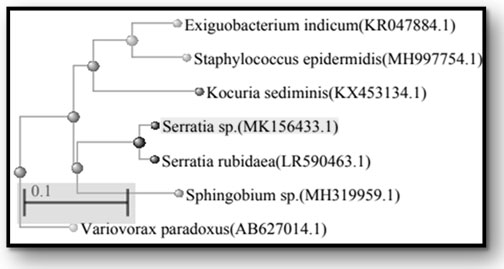
The sequence analysis of 16S rDNA of P1M4, P1M6, P1M7, P1M8, P1M9, P1SM10, and P1SM11 showed the maximum identity of 84 % to Serratia sp. (MK156433.1), 92 % to Exiguobacterium indicum (KR047884.1), 99 % to Kocuria sediminis (KX453134.1), 94% to Variovorax paradoxus (AB627014.1), 95% to Staphylococcus epidermidis (MH997754.1), 99% to Sphingobium yanoikuyae (MH319959.1), and 99% to Serratia rubidaea (LR590463.1), respectively. Results of their closest relatives are shown in phylogenetic tree (Figure 2).
Figure 2: Phylogenetic tree of the identified endophytic bacterial isolates based on the 16S rDNA sequences. The GenBank accession number is given in parentheses for each organism.
Out of 7 bacterial isolates, four isolates (Serratia sp., Variovorax paradoxus, Staphylococcus epidermis, and Serratia rubidaea) showed their ability to solubilize complex calcium phosphate and developed clear zones, ranging from5 to 6.6 mm on Pikovskya’s agar plates (Figure 3). The same isolates also were able tochelate the ironand form yellow zonearound the colony in CAS plate (Figure 4), while threeisolates (E. indicum, K. sediminis, and S. yanoikuyae) had negative results (Table 2).
The enzymatic activity of the bacterial isolates revealed thatall theseisolates produced at least one or otherextracellular enzymes; however, none of the isolates were able to produceall thefivetested enzymes (Table 3). The result showed the maximum a mylolytic activity for Serratia sp., E. indicum, and S. yanoikuyae (Figure5-A) while lipase activity was prominent in S. epidermis followed by Serratia sp., V. paradoxs and S. rubidaea (Figure5-B).
Additionally, all the bacterial isolates had ability to produce protease enzyme (Figure 3-C). Pectinase activity was observed in most of the isolates, S. yanoikuyae, Serratia sp., and E. indicum, K. sediminis and S. rubidaea (Figure5-D).The maximum cellulase activity was observed in S. yanoikuyae, followed by E. Indicum (Figure 3-E).
The quantity of phytohormones varied between the bacterial isolates obtainedfrom H. pterocarpum. The result showed that all the seven isolates producedIAA andGA3, the quantity ranged from 0.002 to 0.056 mg/ml and from 0.006 to 0.144 mg/ml, respectively (Table 4).The IAA data showed that, S. rubidaea produced the maximum amount (0.056 mg/ml), followed byV. Paradoxus and E. indicum with IAA production ranged from 0.12 to 0.13mg/ml. The lowestamounts of IAA (0.002 to 0.007 mg/ml) were obtained for Serratia sp., S. yanoikuyae, K. sediminis and S. epidermis. On the other hand, K. sediminis showed the maximum amount of GA3 (0.144mg/ml), followed by Serratia sp. (0.104mg/ml) while the minimum amounts of GA3were ranged from 0.006 to 0.099 mg/ml.
Figure 3: Production of extracellular enzymes by endophytic bacterial isolates from H. pterocarpum
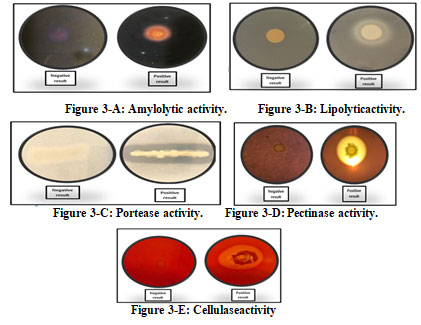
Table 2. Some activity of endophytic bacterial isolated from H. pterocarpum.
| Plate code |
Bacterial isolates |
Biological activities | |
| Phosphate solubilization (mm) | Siderphore production(mm) | ||
| P1 M4 | Serratia sp. | + (6.0) | ++ (16.0) |
| P1 M6 | Exiguobacterium indicum | – | – |
| P1 M7 | Kocuria sediminis | – | – |
| P1 M8 | Variovorax paradoxus | + (5.0) | ++ (16.0) |
| P1 M9 | Staphylococcus epidermis | + (6.6) | ++ (16.0) |
| P1 SM10 | Sphingobium yanoikuyae | – | – |
| P1 SM11 | Serratia rubidaea | + (6.0) | ++ (16.3) |
Note: (+): Positive result, (-): Negative result (no clearing zone).
Table 3. Extracellular hydrolytic enzyme activity of endophytic bacteria isolated from H. pterocarpum.
| Plate code | Bacterial strains | Amylase | Lipase | Protease | Pectinase | Cellulase |
| P1 M4 | Serratia sp. | + | ++ | +++ | +++ | – |
| P1 M6 | Exiguobacterium indicum | + | – | ++++ | +++ | + |
| P1 M7 | Kocuria sediminis | – | – | ++++ | ++ | – |
| P1 M8 | Variovorax paradoxus | – | ++ | +++ | – | – |
| P1 M9 | Staphylococcus epidermis | – | +++ | +++ | – | – |
| P1 SM10 | Sphingobium yanoikuyae | + | – | +++ | ++++ | +++ |
| P1 SM11 | Serratia rubidaea | – | ++ | +++ | + | – |
Note: (-) no clearing zone, (+) weak clearing zone ≤ 5 mm; (++) moderate clearing zone > 5-10 mm; (+++) strong clearing zone ≥ 10-15 mm; and (++++) very strong clearing zone > 15 mm.
Table 4. Production of Indole acetic acid (IAA) and Gibberellic acid (GA3) from endophytic bacteria isolated from H. pterocarpum.
| Plate code | Bacterial isolates | Concentration of IAA (mg/ml) | Concentration of GA3(mg/ml) |
| P1 M4 | Serratia sp. | 0.002±0.01 | 0.104±0.05 |
| P1 M6 | Exiguobacterium indicum | 0.013±0.01 | 0.006 ±0.01 |
| P1 M7 | Kocuria sediminis | 0.006±0.001 | 0.144 ±0.1 |
| P1 M8 | Variovorax paradoxus | 0.012±0.01 | 0.099 ±0.05 |
| P1 M9 | Staphylococcus epidermis | 0.007±0.01 | 0.047±0.4 |
| P1 SM10 | Sphingobium yanoikuyae | 0.002±0.002 | 0.031±0.04 |
| P1 SM11 | Serratia rubidaea | 0.056±0.01 | 0.056±0.01 |
The collected plant was studied and identified as Heliotropium pterocarpum (A. DC.) Hochst. & Steud. ex Bunge. Based on investigation, the beneficial effects of endophytes on plant at extreme environment, diverse bacterial endophytes were isolated from various tissues of H. pterocarpum. It is a very remarkable plant in Saudi Arabia. It is a genus of the flowering plant in the family Boraginaceae, commonly known as heliotropes and has diverse bioactive metabolites including pyrrolizidine alkaloids (Kakar et al., 2010, Radwan and El-shabasy (2020).
Endophytic bacteria inhabitant tissues of H. pterocarpum is relatively unstudied to their endophytic biology and being considered as potential source of novel natural products to be used in industry or agriculture fields.In this study, leaves of H. pterocarpum harbored more endophytic bacteria (5 isolates) compared to stems (2 isolates) and roots (no isolate). This is confirmed with previous studies that have shown leaves of Arabidopsis thaliana harbored more endophytes than roots (Bodenhausen et al., 2013).
As result ofhigh photosynthetic metabolism occurs in the leaf, these products could be utilized by the endophytes. In addition, the most important technique to obtain endophytes from plant is surface sterilization to remove epiphytes on tissues. Hence, a variety of chemical disinfectants have been selected forisolation of bacterial endophytes, however sequential immersion of the plant segments in 70% ethanol and sodium hypochlorite ensured the removal of surface microbial flora (Bacon and Hinton, 1996; Coombs and Franco, 2003).The confirmation of proper surface sterilization of tissues carried out by inoculating last water wash on nutrient agar plate. Absence of any growth after three days incubation indicated the proper surface sterilization of the plant tissues.
The taxonomic status at phylogenetic level of the endophytic bacterial isolates was defined by16S rDNA. The identification of bacterial isolates was recorded and they belonged to Protobacteria (57%), as: (Serratia sp., V. paradoxus, S. yanoikuyae and S. rubidea), Firmicutes (28%) as: (E. indicum and S. epidermis), and Actinobacteria (14%), as: (K. sediminis). In the same regard, endophytic bacteria have been isolated from a number of plant species, as Proteobacteria which is the most predominant phylum, frequently isolated from plants (Afzal et al., 2019). Also, members of Actinobacteria and Firmicutes are the most commonly found as endophytes (Reinhold-Hurek and Hurek, 2011, Fadiji and Babalola, 2020).
The importance of endophytic bacteria is known since long time and they play specific roles in promoting plant growth and protecting the host plants against pathogens and diseases (Muzzamal et al., 2012). Hence, the biological impact of these bacterial strains as plant growth promoting activities were screened for P-solubilization, siderophore production, extracellular enzyme, IAA and GA3 production. In particular, 57% of endophytes isolates were able to solubilize phosphor and produce of siderophor.
Similar research has been documented by Rodríguez et al. (2006) bacterial strains belonging to the genera of Pseudomonas, Bacillus, Rhizobium, Burkholderia, Achromobacter, Agrobacterium, Microccocus, Aerobacter, Flavobacterium, and Erwinia have the ability to solubilize inorganic phosphate (tri-calcium phosphate), (di-calcium phosphate), and rock phosphate.The endophytes isolate which have ability to solubilize phosphorwere able to produce siderophore, in vitro. This can occur by lowering pH by endophytes producing low molecular weight organic acids, in which hydroxyl and carboxyl groups can chelate cations bound to phosphate (Kpomblekoua and Tabatabai, 1994, Fadiji and Babalola, 2020).
The prominence of siderophore for promoting plant growth through attracting the available iron, in therhizosphere was reported.As a result, as, iron be available to plants, but unavailable to phyto-pathogens which could contribute to protect the plant (Pashapour et al., 2016). Malfanova (2013) suggested that endophytes including strains of Pseudomonas fluorescens produce siderophores and can act as biocontrol agents that antagonize growth of some fungal pathogens.
Most of the isolated bacteria in this study were able to produce extracellular enzymes such as amylases, lipases, proteases, pectinase, and cellulase. It can be summarized that extracellular enzymes may play a significant part in mechanism of endophytes colonization into host plant. Also, these enzymescould be included within the attack of plant pathogens in host plant, as reported for Azoarcus sp. (Hurek et al., 1997). Therefore, these bacterial isolates could be used as potential sources of commercial enzyme production for exploitation in medicine, agriculture, and industry (Guo et al. 2008, Fadiji and Babalola, 2020).
Additionally, the isolates produced varied amounts of IAA, and GA3 hormones. These hormones enhance the growth of various plants. The amount of IAA produced by the isolate was increased by the addition of precursor tryptophan in the medium (Uma-Maheswari et al., 2013). The result exhibited the efficiently isolate Serratia rubidaea produced the highest IAA (0.056 mg/ ml). Similarly, Kamilova et al., (2005) reported that Pseudomonas fluorescens– WCS365, stimulated growth of radish root through production of IAA in the presence of tryptophan. Thus, IAA has many different effectsin stimulating elongation and division of plant cell which posterior to growth and development of plant (Phetcharat and Duangpaeng, 2012, Fadiji and Babalola, 2020).
In addition, the bacterial isolates were produced GA3,ranged from 0.006 to 0.144 mg/ml.The GA3 has a role in plant growth, promotes primary and lateral root elongation, and increases yield (Bottini et al., 2004). Report study indicated that B. pumilus isolated from the rhizosphere had the growth promoting effect of red pepper and this effect originated from GA production (Joo et al., 2004).Briefly, beneficialactivities by bacterial endophytes for promotingplant growth are vital factor that could affect plant development in extreme environment.
CONCLUSION
This is probably the first study that demonstrates the diversity of endophytic bacteria in H. pterocarpum, which was collected from Hot spring, Gomyqah, Saudi Arabia. Characterization of these bacterial endophytes includes P-solubilization, siderophore production, extracellular enzymatic activity, phytohormones production was performed in terms of their plant growth-promoting abilities. The successful traits of these bacterial endophyte suggest that they can be utilized in future applications, as biological product through increasing and promoting of plant growth, and protecting plant against pathogens, which help to eliminate or minimize using of commercial fertilizers, and pesticides. Putting all these in consideration, endophytes have a positive impact on plant, environment, and agriculture field.
REFERENCES
Afzal, I., Shinwari, Z. K., Sikandar, S. and Shahzad, S. (2019). Plant beneficial endophytic bacteria: mechanisms, diversity, host range and genetic determinants. Microbiology Research, 221, 36–49.
Alfarhan, A., and Thomas J. (1994). The identification of vascular plant –families in Saudi Arabia. Saudi Biological Society. King Saudi University.
Bacon, C.W., and Hinton D.M. (2002). Endophytic and biological control potential of Bacillus mojavensis and related species. Biological Control. 23: 274–284.
Bodenhausen, N., Horton, M.W. and Bergelson, J. (2013). Bacterial communities associated with the leaves and the roots of Arabidopsis thaliana. Plosone, 8: e56329.
Bottini, R., Cassan, F., and Piccoli, P. (2004). Gibberellins production by bacteria and its involvement in plant growth promotion and yield increase. Applied Microbiology and Biotechnology, 65:497- 503.
Catalogue of Life (2015). Access date March 27, 2019, from: www.catalogueoflife.org.
Chaudhary, S.A. (2001). Flora of the Kingdom of Saudi Arabia, Riyadh: Ministry of Agriculture and Water, Saudi Arabia.
Claus, W.G. (1988). Understanding microbes; a laboratory text book for microbiology. 1st edition. New York (NY): WII Freeman Co.
Coombas, J.T., and Franco, C.M. (2003). Isolation and Identification of Actinobacteria from Surface-Sterilized Wheat Roots. Applied environmental Microbiology, 69: 5603–5608.
Conrath, U., Beckers, G.J.M., Flors, V., Garcia-Agustin, P. et al. (2006). Priming: getting ready for battle. Molecular Plant-Microbe Interactions. 19:1062–1071.
Cotty, P.J., Cleveland, T.E., Brown, R.L.and Mellon, J.E. (1990). Variation in polygalacturonase production among Aspergillus flavus isolates. Applied environmental Microbiology,56: 3885-3887.
El-Naggar, S., El-Hadidy, A., and Olwey, A. (2015).Taxonomic revision of the genus Heliotropium (Boraginaceae) in south Yemen. Nordic. Journal of Botany, 33: 401–413.
Fadiji AE and Babalola OO (2020). Elucidating Mechanisms of Endophytes Used in Plant Protection and Other Bioactivities With Multifunctional Prospects. Front. Bioeng. Biotechnol., 8: 467-471.
Goryluk-Salmonowicz, A., Orzeszko-Rywka, A., Piórek, M., Rekosz-Burlaga, H et al. (2018). Plant growth promoting bacterial endophytes isolated from Polish herbal plants. Acta Scientiarum Polonorum Hortorum Cultus, 17(5), 101–110.
Guo, B., Wang, Y., Sun, X., and Tang, K. (2008). Bioactive natural products from endophytes: a review. Applied Biochemistry Microbiology, 44:136-142.
Govindarajan, M., Kwon, S. and Weon, H. (2007). Isolation, molecular characterization and growth-promoting activities of endophytic sugarcane diazotroph Klebsiella sp. GR9.World Microbial Biotechnology, 23: 997-1006.
Hurek, T., Reinhold-Hurek, B., van Montagu, M. and Kellenberger, E. (1994). Root colonilization and systematic spreading of Azoarcus sp. Strain BH72 in grasses. Journal Bacteriology, 176:1913-1923.
Holbrook, A.A., Edge,W. and Bailey, F. (1961). Spectrophotometric method for determination of gibberellic acid. Advances in Chemistry Series, 28: 159-167.
Hallmann, Q.A., Benhamou, A.N. and Kleopper, J.W. (1997) Bacterial endophytes in cotton: mechanisms of entering the plant. Canadian Journal of Microbiology, 43: 577-582.
Joo, G. J., Kim, Y. M., Lee, I. J., Song, K. S. and Rhee, I. K. (2004). Growth promotion of red pepper plug seedlings and the production of gibberellins by Bacillus cereus, Bacillus macrolides and Bacillus pumilus. Biotechnology Letters. 26:487-491.
Kakar, F., Akbarian, Z.,Leslie, T., Mustafa, M.L.,Watson, J., Hans, P. et al. (2010).An outbreak of hepatic veno-occlusive disease in Western Afghanistan associated with exposure to wheat flour contaminated with pyrrolizidine alkaloids. Journal of Toxicology, 39: 1122-1229.
Kamilova, F., Validov, S., Azarova, T., Mulders, I., Lugtenberg, B. (2005). Enrichment for enhanced competitive plant root tip colonizers selects for a new class of biocontrol bacteria. Environmental Microbiology, 7: 1809-1817.
Kpomblekoua, K., and Tabatabai, M. (1994). Effect of organic acids on release of phosphorus from phosphate rocks. Soil Science, 158(6): 442-449.
Lavakush, J. Y., and Verma,J. P. (2012). Isolation and characterization of effective plant growth promoting rhizobacteria from rice rhizosphere of Indian soil. Asian Journal of Biological Sciences, 5:294-303.
Louden, B.C., Harmann, D., and Lynne, A. M. (2011). Use of blue agar CAS assay for siderophore detection. Journal of Microbiology & Biology Education, 12(1): 51-59.
Malfanova, N., Lugtenberg, B.and Berg,G. (2013). Bacterial endophytes: who and where, and what are they doing there? In: Molecular Microbial Ecology of the Rhizosphere; de Bruijn F J, Ed. ch 36, Wiley- Blackwell, Hoboken, NJ, USA, pp. 393-403.
Miliute, I., Buzaiteodeta, B. D., and Stanys, V. (2015). Bacterial endophytes in agricultural crops and their role in stress tolerance: a review. Zemdirbyste-Agriculture, 102 (4): 465–478.
Muzzamal, H., Sarwar, R., Sajid, I. and Hasnain, S.H. (2012). Isolation, identification and screening of endophytic bacteria antagonistic to biofilm formers. Zoological Society of Pakistan, 44(1): 249-257.
Nair, D. N. and Padmavathy, S. (2014). Impact of endophytic microorganisms on plants, environment and humans. The Scientific World Journal, (2):250-253.
Ngoma, L., Esau, B., and Babalola, O.O. (2013). Isolation and characterization of beneficial indigenous endophytic bacteria for plant growth promoting activity in Molelwane Farm, Mafikeng, South Africa. African Journal of Biotechnology, 12(26): 4105-4114.
Pashapour, S., Besharati, H., Rezazade, M., Alimadadi, A., and Ebrahimi, N. (2016). Activity screening of plant growth promoting rhizobacteria isolated from alfalfa rhizopshere. Biological Journal of Microorganism, 4(16): 122-129.
Patten, CL., and Glick, BR. (2002). Role of Pseudomonas putida indole acetic acid in development of the host plant root system. Applied Environmental Microbiology, 68:3795–3801.
Patten, D. M., and Glick, B.R. (2003). Methods for isolating and characterizing ACC deaminase‐containing plant growth‐promoting rhizobacteria. Physiologia Plantarum, 118(1): 10-15.
Phetcharat, A., and Duangpaeng, A. (2012). Screening of endophytic bacteria from organic rice tissue for indole acetic acid production. Procedia Engineering, 32:177 – 183.
Radwan, D. M. and El-shabasy, A. E. (2020). Comparative Analysis of Five Heliotropium species in Phenotypic Correlations, Biochemical Constituents and Antioxidant Properties. CATRINA, 21(1): 1-8.
Rajan, A., Kumar, D.S. and Nair, A.J. (2011). Isolation of a novel alkaline lipase producing fungus Aspergillus fumigatus MTCC 9657 from aged and crude rice bran oil and quantification by HPTLC. International Journal of Biological Chemistry, 5: 116-126.
Reinhold-Hurek, B., and Hurek, T. (2011). Living inside plants: bacterial endophytes, Current Opinion in Plant Biology, August 2011, Pages 435-443., 14 (2011), pp. 435-443
Rodriguez, H., Fraga, R., Gonzalez, T.and Bashan, T. (2006). Genetic of phosphate solubilisation and its potential applications for improving plant growth-promoting bacteria. Plant Soil. 287:15-21.
Santoyo, G., Moreno-Hagelsieb, G., Orozco-Mosqueda, M., Glick, B.R. (2016). Plant growth-promoting bacterial endophytes, Microbiological Research, Volume 183:92-99.
Schwyn, B., and Neilands, J. B. (1987).Universal chemical assay for the detection and determination of siderophores. Analytical Biochemistry, 160:47–56.
Singh, G., Singh, N.andMarwaha TS.(2009). Crop genotype and a novel symbiotic fungus influences the root endophytic colonization potential of plant growth promoting rhizobacteria. Physiology and Molecular Biology of Plants. 15:87-92.
Thomas, J., (2011). Flora of Saudi Arabia, checklist, 2011. Access date, 25 September 2019,from:www.plantdiversityofsaudiarabia.info/Biodiversity-SaudiArabia/Flora/Checklist/Cheklist.htm
Tennalli, G., Udapudi, B.andNaik, P. (2012) Isolation of proteolytic bacteria and characterization of their proteolytic activity. International Journal of Advanced Science and Technology;2: 185-192.
Uma-Maheswari, T., Anbukkarasi, K., Hemalatha, T. and Chendrayan, K.(2013). Studies on phytohormone producing ability of indigenous endophytic bacteria isolated from tropical legume crops. International Journal of Current Microbiology and Applied Science; 2(6): 127-136
Wilson, D. (1995). Endophyte: the evolution of a term, and clarification of its use and definition. Oikos: 274-276.
Zaghloul, R.A., Abou-Aly, H.E.,Tewfike, T.A.andAshry, N.M. (2016).Isolation and characterization of endophytic bacteria isolated from legumes and non-legumes plants in Egypt. Journal of Pure and Applied Microbiology; 10: 277-290.
Zinniel, D.K,Lambrecht, P., Harris, N.B., Feng, Z.andKuczmarski, D. (2003).Isolation and characterization of endophytic colonizing bacteria from agronomic crops and prairie plants. Applied and Environmental Microbiology; 68: 2198-2208.


