National Reference Laboratory, Department of Microbiology, Bhopal Memorial Hospital and Research Centre (BMHRC), Bhopal.M.P.
Article Publishing History
Received: 12/06/2019
Accepted After Revision: 15/09/2019
Tuberculosis remains a major cause of morbidity & mortality globally. Rapid diagnosis of MTB is important for prevention of tuberculosis. Sputum smear microscopy does not differentiate between viable and dead bacilli. Ribosomal RNA (rRNA) based methods are one of the important tools for rapid detection of viable MTB from patients samples. The aim of the study was to detect MTB in Sputum samples of follow up patients of MTB by Reverse Transcriptase RT PCR and to analyze the results of RT PCR with smear microscopy. 211 follow up sputum Samples were received through the Revised National Tuberculosis Control Programme (RNTCP). RNA was extracted from culture isolates and then processed by Reverse Transcriptase RT PCR targeting 16SrRNA gene. Direct smear microscopy of all sputa were done prior to processing. The RT PCR assay showed overall 59.87% accuracy. Out of a total 211 samples, 66 (31.2 %) were positive and 145 (68.7) were negative for Reverse transcriptase RT PCR. Of 66 RT positive samples, 38 (57.5 %) were smear positive and 28 (42.4 %) were smear negative. Of 145 RT negative samples, 33 (15.6%) were smear positive and 112 (53%) were smear negative.RT PCR could detect viable MTB in smear negative samples with 57.88% sensitivity (CI 95%, 44.79% to 69.66%) and with 77.24% specificity (69.55% to 83.79%).To conclude,Reverse transcriptase RT PCR may prove to be a promising tool for early detection of Mycobacterium tuberculosis.
Follow up,MTB, Smear Microscopy, Reverse Transcriptase, RT PCR, 16SrRNA.
Pillai A, Panwalkar N, Desikan P. Reverse Transcriptase Polymerase Chain Reaction: A Promising tool for Rapid Identification of Mycobacterium tuberculosis. Biosc.Biotech.Res.Comm. 2019;12(3).
Pillai A, Panwalkar N, Desikan P. Reverse Transcriptase Polymerase Chain Reaction: A Promising tool for Rapid Identification of Mycobacterium tuberculosis. Biosc.Biotech.Res.Comm. 2019;12(3). Available from: https://bit.ly/2m2MPmJ
Copyright © Pillai et al., This is an open access article distributed under the terms of the Creative Commons Attribution License (CC-BY) https://creativecommns.org/licenses/by/4.0/, which permits unrestricted use distribution and reproduction in any medium, provide the original author and source are credited.
Introduction
Tuberculosis (TB) remains a major cause of morbidity and mortality globally. Rapid diagnosis of TB is vital for prevention of further transmission of tuberculosis (PMDT 2017). Smear microscopy is a rapid, simple and inexpensive technique which is highly specific for the diagnosis of active tuberculosis. However, sputum smear microscopy has considerable limitations. The sensitivity is grossly compromised when the bacterial load is less than 10,000 organisms/ml sputum sample. Moreover, it does not differentiate between viable and dead bacilli (Honeyborne et.al 2011) . Ribosomal RNA (rRNA) based methods are one of the important tools for rapid detection of viable MTB from patients samples. rRNA constitutes 80% of total RNA and is the most conserved region which is structurally more stable and have longer half – life, (Belasco et.al 1986, Desikan 2013, PMDT 2017 and WHO 2018). In the present study, in addition to the smear microscopy, rRNA based reverse transcriptase real time PCR targeting 16SrRNA gene has been used as a technique for rapid detection of Mycobacterium tuberculosis.
Materials and Methods
A total of 211 sputum samples previously diagnosed as pulmonary tuberculosis were examined in the study. The study was approved by Institutional Ethics Committee (IEC), BMHRC. Smears of all sputum samples were prepared and stained with Ziehl –Neelsen stain to observe the presence of acid fast bacilli by bright field microscopy under the 100X objective. Grading of smears was performed as per the criteria defined by RNTCP (RNTCP 2018). The sputum samples were digested and decontaminated by N-acetyl-L cysteine-sodium hydroxide-Citrate method as per the guidelines by RNTCP (RNTCP 2018).
The sputum samples were cultured on LJ medium since culture is a gold standard. Extraction of RNA from the decontaminated sediments was performed by a commercially available RNA extraction kit (Nucleopore RNA isolation kit, Genetix Biotech Asia Pvt.ltd) as per the manufacturer’s protocol. The synthesis of cDNA was carried out with a commercially available kit (High Capacity cDNA reverse transcription kit) as per the manufacturer’s protocol . In brief, reaction mixture was prepared by adding 10x RT PCR Buffer-2ul, 25x Dntp Mix-0.8ul, 10x RT Random Primer-2ul, Multiscribe reverse transcriptase-1ul, RNAase Inhibitor-1ul, Nuclease Free Water was added to make the final reaction volume of 10ul. The PCR was carried out with the following cycling conditions: 370C for 120 minutes and then 850C for 5 minutes for 45 cycles.
Real Time PCR
The detection of MTB using 16SrRNA gene in sputum samples was analyzed by Light Cycler 2.0 (Roche Diagnostics, Meylan, France) using a commercially available kit (fast start Essential DNA Probe master mix, Roche Diagnostics, Meylan, France) . Primers and probes were synthesized commercially as published in the previous literature (Juan et.al 2012) MTBC 16S forward (5′-GGGATGCATGTCTTGTGGTG-3′) and MTBC 16S reverse (5′-CCGTCGTCGCCTTGGTAG-3′) primers, which amplify 100 bp fragment of the 16SrRNA gene, and a 21 bp Taqman probe (5′-CGGGCTCATCCC ACACCGCTA-3′) labeled at the 5′ end with 6-carboxyfluorescein (6-FAM), and at the 3′ end with the quencher N,N,N,N-tetramethyl-6-carboxyrhodamine (TAMRA) (Invitrogen, Carlsbad, CA, USA) were used for RT PCR. The master mix was prepared by adding probe master-10ul, Forward primer-1ul, Reverse primer-1ul, Probe-1.6ul and Nuclease Free Water to make the final volume of 15ul. The Real Time PCR was run with the following cycling conditions, at 500C for 2 min to denature the DNA template, 950C for 10 min to activate the Taq polymerase, followed by 40 cycles at 950C for 15s.
Results and Discussion
Out of a total of 211 samples,140 (66.3%) were smear negative and 71 (33.6%) were smear Positive. Out of 211 samples, 66 (31.2 %) were positive and 145 (68.7%) were negative for MTB by reverse transcriptase RT PCR. Of 66 RT PCR positive samples, 38 (57.5 %) were smear positive and 28 (42.4 %) were smear negative. Of 145 RT PCR negative samples, 33 (22.7%) were smear positive and 112 (77.2%) were smear negative (Table-1). In our study 16S rRNA based RT PCR had 57.88% sensitivity (CI 95%, 44.79% to 69.66%) 77.24% specificity (CI 95%, 69.55% to 83.79%), and overall 71.09% accuracy (CI 95%, 64.47% to 77.11%) for detection of MTB.(Table 1,Figure 1 and table 2). However smear microscopy had 49.30% sensitivity (CI 95% , 37.22% to 61.44%), 84.29% specificity (CI 95%, 77.18% to 89.88%),PPV 61.40% ( CI 95%, 50.35% to 71.40%), NPV 76.62% (72.05% to 80.65%) and overall 72.51% accuracy.While both tests are rapid tests, performance characteristics of16S rRNA based real time PCR are significantly higher than that of smear microscopy.
In our study it was found that there were 3 samples which were culture positive but RT PCR negative.This may be due to the presence of PCR inhibitors in the sputum samples, mismatch of primer pairing or the presence of fragmented tubercle bacilli that might have resulted in suboptimal quality of RNA.The sensitivity of rRNA based RT PCR was found to be more over the culture in our study, indicated by 12 samples that were culture negative but RT PCR positive. Given the high sensitivity and specificity, molecular assays are widely accepted as a promising diagnostic tool for the detection of MTB (Woese 1987). rRNA constitutes 80% of the total RNA and is structurally more stable than messenger RNA(mRNA). Moreover, rRNA is present at 1000 – 10000 times more in copy numbers than genomic DNA, therefore, rRNA, particularly 16S rRNA, is a good target for the detection of MTB, (Woese 1987) from the clinical samples.
The detection of 16S rRNA is a reflection of the metabolic state of the total population of bacteria, therefore, the measurement of 16S rRNA can be used as an indicator of viability (Hellyer et.al 1999).The average turnaround time of our study was found to be three days. 16SrRNA based real time PCR therefore appears to be a promising tool for rapid identification of Mycobacterium tuberculosis. The challenge would be to make 16S rRNA based Real Time PCR available and accessible to the population that needs TB diagnosis but can afford it to least. Further studies and innovations to make it a cost effective test are the need of the hour.
Table 1: RT PCR and Microscopy (n=211)
| RT PCR Positive | RT PCR Negative | Total | ||||
| No. | % | No. | % | No. | % | |
| Smear Positive | 38 | 57.5 | 33 | 22.7 | 71 | 33.6 |
| Smear Negative | 28 | 42.4 | 112 | 77.2 | 140 | 66.35 |
| Total | 66 | 31.2 | 145 | 68.7 | 211 | |
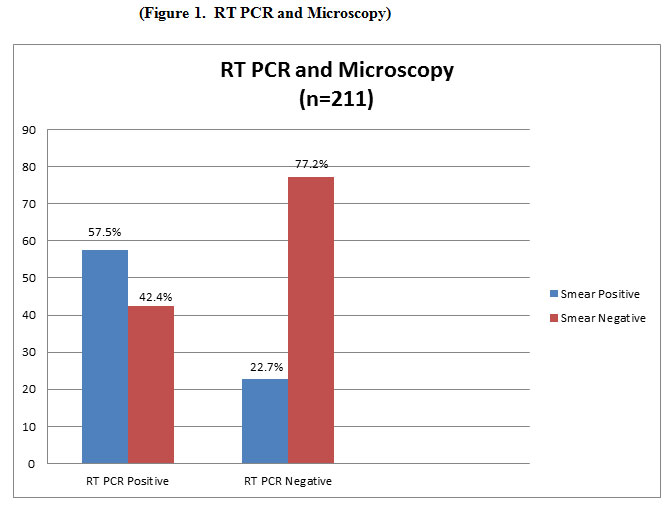 |
Figure 1: RT PCR and Microscopy |
Table 2: Performance parameters of RT PCR with respect to Smear Microscopy
| Method | Result | RT PCR Positive (n=211) | RT PCR
Negative (n=211) |
Sensitivity
(%) |
Specificity
(%) |
Positive
Predictive value (%) |
Negative
Predictive value (%) |
Accuracy
(%) |
| Smear Microscopy | Positive
Negative |
38
28 |
33
112 |
57.5 | 77.2 | 53.5 | 80 | 71.09 |
References
Belasco, J. G., G. Nilsson, A. von Gabain, and S. N. Cohen. (1986) The stability of E. coli gene transcripts is dependent on determinants localized to specific mRNA segments Cell Vol.46: Pages 245–251
Desikan P. (2013) Sputum smear microscopy in tuberculosis: Is it still relevant? Indian Journal of Medical Research. Vol 137 No 3 Pages : 442–444
Hellyer, L. E. Desjardin, G. L. Hehman, M. D. Cave, K. D Eisenachi (1999) Quantitative Analysis of mRNA as a Marker for Viability of Mycobacterium tuberculosis,Journal of Clinical Microbiology Vol 37 No 2: Pages 290–295
Honeyborne I, Timothy D. McHugh, Patrick P. J. Phillips, Selina Bannoo, (2011) Molecular Bacterial Load Assay, a Culture-Free Biomarker for Rapid and Accurate Quantification of Sputum Mycobacterium tuberculosis Bacillary Load during Treatment. Journal of clinical microbiology, Vol. 49, No. 11: Pages 0095-1137
Jiang, Li Juan, Wen Juan Wu, Hai Wu, Son Sik Ryang, Jian Zhou (2012) Rapid Detection and Monitoring Therapeutic Efficacy of Mycobacterium tuberculosis Complex Using a Novel Real-Time Assay. Journal of Microbiology and Biotechnology.Vol.22 No 9: Pages 1301–1306
PMDT (2017) Programmatic Management of Drug Resistant Tuberculosis (PMDT) Guidelines (2017)
Revised National Tuberculosis Control Programme (RNTCP) (2018) Dots Plus Guidelines, Central TB Division, Directorate General of Health Services,Ministry of Health and family welfare,Nirman Bhawan New Delhi https://tbcindia.gov.in. (2018)
Woese C R. (1987) Bacterial evolution Microbiological Reviews Vol 51 Pages: 221– 271.
World Health Organization (2018) (WHO) Global Tuberculosis report (2018) http;//www.who.int/tb/publications/global-report/en/.


