Seri-Biotech Research Laboratory, Central Silk Board, Carmelaram Post, Kodathi, Bangalore, 560 035, India.
Corresponding author Email: ravikumarpillai@gmail.com
Article Publishing History
Received: 21/10/2017
Accepted After Revision: 18/12/2017
Microsporidia are obligate intracellular, spore-forming parasites that infect both invertebrates and vertebrates. They infect silkworms causing the deadly pebrine disease leading to heavy crop loss in sericulture. Because of the horizontal and vertical transmittance, outbreaks should be detected at an early stage and persistent infections should also be identified to prevent further transmittance. So far, microscopic examination method remains the conventional detection method for screening of microsporidia in sericulture. Molecular diagnosis tools have an advantage over microscopic detection as they are more specific, sensitive and aid in early detection. Microsporidia detection by PCR method using primers designed from SSU-rRNA is widely used. In this study, we developed a PCR assay for the detection of microsporidia using primers designed from the conserved regions of RNA polymerase gene. Under optimized PCR conditions, the assay yielded a ~650 bp DNA fragment from microsporidia infected silkworms, Bombyx mori and Antheraea mylitta. Sequence analysis of the amplified products has shown homology to various microsporidia including Nosema bombycis and N. antheraea. No non-specific products were observed. This method could help in early detection of microsporidia infection at any developmental stage of the silkworm and thereby reducing the crop loss.
Detection, Microsporidia, Pcr
Roy G, Mandal K, Ravikumar G. PCR-Based Detection of Microsporidia in Silkworms Using Non-Conventional RNA Polymerase Primers. Biosc.Biotech.Res.Comm. 2017;10(4).
Roy G, Mandal K, Ravikumar G. PCR-Based Detection of Microsporidia in Silkworms Using Non-Conventional RNA Polymerase Primers. Biosc.Biotech.Res.Comm. 2017;10(4). Available from: https://bit.ly/2WBvsuO
Introduction
Silkworm, Bombyx mori is one of the most important domesticated insects, which produces luxuriant silk thread in the form of cocoon by consuming mulberry leaves during larval period. In India the bulk of the commercial silk produced is mulberry silk whereas, Eri, Tasar & Muga silk contribute to a lesser extent. These silkworms are susceptible to various diseases resulting in substantial crop loss which is estimated to be 40% in India (Singh et al., 2012). The common pathogens infecting them are microsporidians including Nosema
bombycis, nucleopolyhedrovirus (NPV) and densovirus (mainly DNV1&2), infectious flacherie virus (IFV), cytoplasmic polyhedrovirus (CPV) and bacteria. The microsporidians cause pebrine disease; NPV causes grasserie; and DNVs, IFV and bacterial pathogens cause flacherie diseases. Among all, the microsporidians disease is responsible for the significant economic loss in the sericulture industry. Microsporidiasis remained a threat to silk industry since time immemorial, because of its unique and recurrent occurrence and is the only disease transmitted both horizontally and vertically (Bhat et al., 2009). Several species and strains of microsporidia have been isolated from infected silkworms among which pebrine caused by Nosema bombycis is the most prevalent. Other microsporidian species (Vairimorpha, Pleistophora, Thelohania etc.) which differ in their spore morphology, sites of infection and virulence, have also been isolated from silkworms (Kawarabata, 2003, Gupta et al., 2017).
Since the control of disease is often met with limited success, early detection of pathogens is essential to control of emerging, reemerging, and in preventing the spread of infectious diseases. Microsporidian are easily detected by light microscopy when infections are heavy and spores are present. However, early infections without spores, or light infections with low numbers of spores are easily missed. This limitation has made it difficult to conduct investigations into microsporidian prevalence and transmission. To overcome these difficulties, PCR- based techniques have been developed to detect the major pathogens of silkworms with great specificity and sensitivity (Hatakeyama and Hayasaka 2003, Hamiduzzaman et al., 2010, Ravikumar et al., 2011, Fu et al., 2016).
Due to the availability of sequence information and the presence of conserved and variable sequence regions within the SSU rRNA genes, PCR-based methods have typically used primers of this gene for the detection of microsporidians (Franzen and Muller, 1999). Herein, we report that primers designed from the RNA polymerase of microsporidians can also be used to detect microsporidians from silkworms. To our knowledge, this is the first report on the detection of microspordians using its RNA polymerase primers from silkworms.
Materials and Methods
Silkworm and Microsporidian Infection
The silkworm rearing and microsporidian infection were essentially performed as reported by us (Ravikumar
et al, 2011). Silkworms, B. mori (Pure Mysore) were fed on mulberry leaves. For Microsporidian infection, 3rd instar day 1 larvae were orally fed with 2000 spores/larva and periodical observations were taken. Control larvae did not receive microsporidian infection. On 4th and 8th day post infection (p.i.), larval mid gut tissues were dissected out and used for DNA extraction, followed by PCR. DNA was also extracted from pupa, adult and eggs of infected and normal silkworms.
Dna Extraction
DNA extracted from the mid gut of infected and control using Hi-Pure DNA extraction Kit (Himedia) according to manufacturer’s protocol. DNA from mulberry leaves and pebrine infected A. mylitta DNA were used as reported earlier (Ravikumar et al., 2011). The DNA was analyzed in 1% agarose gel electrophoresis and quantified using a Nanodrop (Thermo Corporation) spectrophotometer.
Pcr and Cloning
A set of primers were designed from the conserved region of available microsporidian RNA polymerase sequences from NCBI database. The primers used were: Sense: 5’-CCICAYTTYCCIAARGARGAYTA-3’ and antisense: 5’-AARGAYITIGARGGIACIAAYGA-3’. (I: deoxyinosine; R: A, G; Y: T, C). PCR reactions were carried out using 1X Taq buffer, 2.5 mM dNTPs, 25 mM MgCl2, 0.5U Taq DNA polymerase (Fermentas) and 100 ng of DNA. The DNA from control silkworms and mulberry DNA were employed as negative controls. PCR reactions were carried out (Eppendorf) using the following cycles: 94°C for 2 min, 30 cycles of 94°C for 40 s, 48°C for 30 s, and 72°C for 30s and 1 cycle of 72° C for 5 min. PCR products were analyzed in 1% agarose gel electrophoresis, stained in Sybergreen (HiMedia) and visualized under UV transillumination. The PCR products were cloned in pJET blunt end cloning vector (Fermentas) and positive clones were confirmed by colony PCR. Purified plasmids were sequenced at Eurofins, Bangalore, followed by BLAST analysis (NCBI).
Results and Discussion
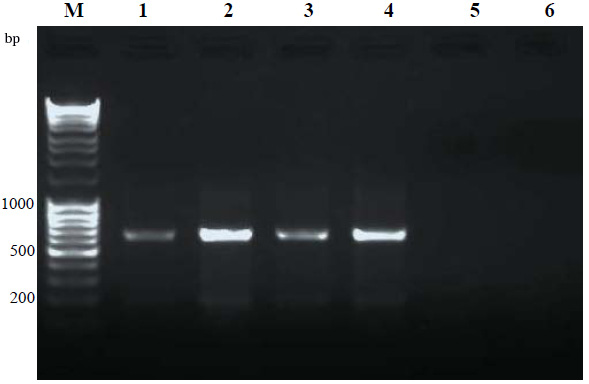
Results are presented in Figure 1. PCR amplifications have resulted in discrete and desired product. DNA extracted from the microsporidian infected silkworm yielded specific amplification products of ~ 650 bp (Lane 1-4) using RNA polymerase primers. No non- specific products were observed. The negative controls; DNA from normal silkworm and the plant DNA from mulberry showed no PCR products, indicating the specificity of the PCR. The banding intensity on day 8th was higher to that of on 4th day showing the proliferation of pathogen at an advanced stage of infection. Further confirmation of the PCR products was done by sequencing and BLAST analysis. BLAST showed 92- 99 % homology to RNA polymerases of various isolates of N. bombycis, Nosema Sp, N. antheraea, N. ceranae, N. pernyi and other microsporidia. The same results were obtained from other developmental stages, pupa, adult and eggs (data not shown) of the silkworm. In addition, the same primer sets could detect microsporidian of tasar silkworm A. mylittashowing the efficacy of the RNA polymerase-based primers in detecting microsporidian of other silkworm species than B. mori. The conserved regions of RNA polymerase gene was effectively utilized for the detection of microsporidia in the present work and it can be used for detection of microsporidia of other insects/organisms also. Highly conserved SSU-r RNA gene primers were successfully used for the detection and classification of microsporidia across organisms with high specificity and sensitivity (Weiss and Vossbrinck, 1999; Jehle et al., 2006, Ravikumar et al., 2011). PCR diagnosis of N. pernyi using SSU-rRNA primers provided increased specificity and sensitivity when compared with light microscopy in Antheraea pernyi (Jiang et al., 2011). For the effective control of pebrine disease, outbreaks should be detected at an early stage and persistent infections should also be identified to prevent further transmittance of the disease.
In our study, microsporidia were detected by PCR at 4th day of p.i, whereas the spores were visible under microscope only on day 8 and afterwards. Hence, this method can be useful in the early detection of microsporidia which is critical in reducing crop loss in sericulture. Further, real-time quantitative PCR assay can be used with RNA polymerase primers for increased sensitivity. The results of this study suggest that RNA polymerase primers from microsporidia can be used for pebrine detection in sericulture. To the best of our knowledge, this is the first study in which PCR was used for the successful detection of microsporidia using RNA polymerase primers in silkworms.
Acknowledgements
Authors are thankful to Messrs. S. N. Gundurao and N. Pillapa for rearing silkworms. Financial support was provided by Central Silk Board (CSB), Bangalore, India. Mr. Gourab Roy is thankful to CSB for a Junior Research Fellowship.
References
- Bhat S. A., Bashir I., Kamili A. S. (2009) Microsporidiosis of silkworm, Bombyx mori(Lepidoptera- Bombycidae): A review, African Journal of Agricultural Research, 4, 1519-1523.
- Franzen C. and Müller A. (1999) Molecular techniques for detection, species differentiation and phylogenetic analysis of microsporidia, Clinical Microbiology Reviews, 12, 243-285.
- Fu Z., He X., Cai S., Liu H., He X., Li M., Lu X. (2016) Quantitative PCR for detection of Nosema bombycisin single silkworm eggs and newly hatched larvae Journal of Microbiological Methods, 120, 72-78.
- Gupta S. K., Hossain Z., Nanu M. M., Mondal K. (2017) Impact of microsporidian infection on growth and development of silkworm Bombyx mori(Lepidoptera: Bombycidae), Agriculture and Natural Resources, 50, 388-395.
- Hamiduzzaman M., Novoa E. G., Goodwin P.H. (2010) A multiplex PCR assay to diagnose and quantify Nosemainfections in honey bees (Apis mellifera), Journal of Invertebrate Pathology, 105, 151-155.
- Hatakeyama Y. and Hayasaka S. (2003) A new method of pebrine inspection of silkworm egg using multiprimer PCR, Journal of Invertebrate Pathology, 82, 148-151.
- Jehle J.A., Lange M., Wang H., Hu Z., Wang Y., Hauschild R. (2006) Molecular identification and phylogenetic analysis of baculoviruses from Lepidoptera, Virology, 346, 180-193.
- Jiang Y. R., Deng Z. H., Shi S.L., Yang R. S., Li Y.Z., Duan Y. X., Qin L. (2011) Development of a PCR-based method for detection of Nosema pernyi, African Journal of Microbiology Research, 5, 4065-4070.
- Kawarabata T. (2003) Biology of microsporidians infecting silkworm, Bombyx mori, in Japan-Review, Journal of Insect Biochemistry and Sericology, 72, 1-32.
- Ravikumar G., Raje Urs S., Vijaya Prakash B., Rao C.G.P., Vardhana K.V. (2011) Development of a multiplex polymerase chain reaction for the simultaneous detection of microsporidians, nucleopolyhedrovirus, and densovirus infecting silkworms, Journal of Invertebrate Pathology, 107, 193-197.
- Singh T., Bhat M. M., Khan M. A. (2012) Microsporidiosis in the silkworm, Bombyx mori(Lepidoptera: Bombycidae), Pertanika Journal of Tropical Agriculture Science, 35, 387-
- Weiss L. M. and Vossbrinck C. R. (1999) Molecular biology, molecular phylogeny, and molecular diagnostic approaches to the microsporidia. In: Wittner, M., Weiss, L.M. (Eds.), The Microsporidia and Microsporidiosis. American Society for Microbiology, Washington, DC, pp. 129-171.