1, 2Department of Microbiology, Skyline University Nigeria
3Wuhan Botanical Garden, University of Chinese Academy of Sciences, China P. R
4Department of Microbiology, University of Gothenburg, Sweden
Corresponding author email: mustapha.abdulsalam@sun.edu.ng
Article Publishing History
Received: 18/07/2022
Accepted After Revision: 25/09/2022
The goal of this investigation was to see if it was possible to produce α-amylase from agricultural waste (groundnut shell). The strain (M1) identified as Aspergillus sp. exhibited the largest clearance zone (1.6 cm) and was used in fermentation studies. The activity of α-amylase increased after 24 hours of fermentation, peaked at 72.3 U/mL on day 5, and then began to decline. The effect of optimized environmental conditions studied using OFAT, and it was discovered that pH 6, inoculum size of 1 × 107 spores/mL, incubation period of 120 h, substrate concentration of 3 percent (w/v), and temperature of 35oC were the best for producing α-amylase from groundnut shell using Aspergillus sp. In a single fermentation, these optimum conditions were used, and the experiment yielded an optimum enzyme yield of 121.3 U/mL. This research shows that groundnut shell, a low-cost and commonly available waste, could be an ideal substrate for the manufacture of value-added products.
Amylase, OFAT, Fermentation, Optimization, Aspergillus sp.
Abdulsalam M, Fari H.I, Tiamiyu B. B, Salam O. L. Optimizing α-amylase Production from locally Isolated Aspergillus species Using Selected Agro Waste as Substrate. Biosc.Biotech.Res.Comm. 2022;15(3).
Abdulsalam M, Fari H.I, Tiamiyu B. B, Salam O. L. Optimizing α-amylase Production from locally Isolated Aspergillus
species Using Selected Agro Waste as Substrate. Biosc.Biotech.Res.Comm. 2022;15(3). Available from: <a href=”https://bit.ly/3LvuLJo“>https://bit.ly/3LvuLJo</a>
INTRODUCTION
Groundnut (Arachis hypogea.L) originated in Latin America and was introduced to West Africa by Portuguese traders in the 16th century. Developing nations account for 97% of acreage and 94% of worldwide crop production. (Tela et al. 2021). Nigeria was the third largest producer of groundnuts in the world in 2021, trailing only China and India, with output values of 16,685,915, 6,857,000, and 3,028,571 tonnes respectively (Shanthala et al. 2022).
Several researchers have stated that cultivating microorganisms on lignocellulosic materials is a promising strategy for creating enzyme, which will reduce the cost of producing enzyme. Lignocellulosic materials are abundant in the environment, accounting for half of all terrestrial biomass (Sabino et al. 2021) Lignocellulosic wastes are a complex structure made up primarily of cellulose, hemicellulose, and lignin, which are linked by covalent bonds to create a complex network resistant to microbial invasion.
Lignocellulosic wastes are a low-cost source of enzyme production (Sabino et al. 2021). To keep lignocellulosic waste from becoming a nuisance in the environment, it has been used to make a variety of value-added goods, such as enzymes (Santana et al. 2021). Agricultural and industrial operations produce lignocellulosic waste. Sugarcane bark, bagasse and straw, rice straw and rice bran, cassava peel, maize cobs and straw, wheat chaff and bran, banana straw, cassava peel, wood scraps, and groundnut shell are only a few examples of lignocellulosic wastes produced in Nigeria (Igbokwe et al. 2022). It may interest you to learn that these plentiful wastes are underutilized, contributing to pollution problems in the environment. Bioethanol, enzymes, organic acids, biosurfactants, biogas, biohydrogen, and biofertilizers have all been developed using lignocellulosic wastes as a viable substrate due to their high nutrient content (Chen et al. 2022).
Amylases are used to hydrolyze polysaccharides like starch into simple sugar constituents in the starch processing industry. As new opportunities in biotechnology have emerged, the range of amylase applications has expanded to include analytical chemistry, medical and pharmaceutical applications. (Almulaiky, et al. 2021). Amylases are one of the most essential enzymes and are very important in biotechnology; they are a class of industrial enzymes that account for around one quarter of the global enzyme market (Kalia et al. 2021). The key benefit of using microorganisms to produce amylases is the least expensive of mass production as well as the ease with which microbes can be influenced to produce desired enzymes.
However, the cost of generating α-amylase is so expensive, there is a need to develop more cost-effective methods of producing the enzyme. This can be accomplished by making use of widely available and abundant wastes, such as groundnut shell, which is a common solid waste in underdeveloped countries. Its ability to produce α-amylase will facilitate waste management at a low cost, minimize pollution caused by garbage, and expand the country’s economic basis. The goal of this study is to produce α-amylase from cheap and readily available waste (groundnut shell) by optimizing the fermentation conditions by One Factor at a Time (OFAT). The enzyme will be produced by locally isolated Aspergillus sp. isolated from soil samples.
MATERIAL AND METHODS
Groundnut shells were collected from local farmers in Kano State, Nigeria, then crushed into fine powder in a milling machine. The glassware which include test tubes, MacCartney bottle, beakers, conical flask and measuring cylinder were washed with detergent and rinsed thoroughly with water. They were allowed to dry, wrapped in aluminum foil and then sterilized at temperature of 160°C for 60 minutes. The inoculating loop and cork-borer were sterilized by dipping in flame until they were red hot. The spatula was sterilized using alcohol and bent glass rod was sterilized with alcohol and flame (Pasin et al. 2020).
Potato Dextrose Agar was prepared following the manufacturer’s specification while starch agar was prepared following the method described by (Saha and Mazumdar, 2019). The media were sterilized at 121oC for 15 minutes. Soil samples were used to isolate amylase-producing fungus. The soil samples were taken at a landfill site for cassava garbage. Before plating on Potato Dextrose Agar using the pour plate technique, the samples were serially diluted to a concentration of 10-4. The fungal plates were incubated for 3-7 days at 30°C. Isolates with unique clusters were carefully selected after incubation and sub-cultured on fresh media to obtain pure culture. At 4oC, pure cultures were kept in agar slants (Pasin et al. 2020).
All fungi isolates were identified conventionally. Microscopic and macroscopic views of the isolates were used for identification. The colonial morphology of the fungal isolates on the plates was used to identify them. Cultural and morphological factors such as the nature of the hyphae, color of the colonies, appearance of the colonies and growth rates were taken into account for proper characterization of the isolates, as described by (Ani et al. 2021). This involved using a sterile needle to pick a small bit of mycelial mat and placing it on a clean glass slide, staining with lacto-phenol cotton-blue, and covering with a cover slip. There were reproductive and vegetative structures found. During microscopy, the type of spores, sporangia, hyphae branching, and the presence of septa were all considered. A fungal atlas was used to identify the isolates (Ani et al. 2021).
The best isolate used for the production of fungal amylase was being identified at Centre for Biotechnology Research Kano, Kano, Nigeria. The methods used were the sequencing of ITS1 and ITS2 regions of the genomic DNA, followed by comparison of sequence similarity with other fungi on the National Centre for Biotechnology Information (NCBI) (https://blast.ncbi.nlm.nih.gov/Blast.cgi?PAGE_TYPE=BlastSearch). Screening was performed on the isolated organisms using the method outlined by (Saha and Mazumdar 2019). Peptone, 0.90 g/L; (NH4)2HPO4, 0.40 g/L; KCl, 0.10 g/L; MgSO4,7H2O, 0.10 g/L; starch soluble, 10 g/L and 2 percent (w/v) agar-agar were included in the amylase agar. The isolates were streaked on amylase agar and cultured for 7 days at 30oC. After incubation, the plates were filled with iodine solution and incubated for another 30 minutes. After that, the plates were rinsed with double distilled water and looked for a starch hydrolysis zone of clearance around the colony expansion. As amylase producers, microbial colonies with the highest zone of clearance were chosen.
Using an inoculating loop, spores from 7-day-old fungal cultures were scraped and aseptically transferred to sterile distilled water comprising 0.1 percent Tween-80 as described by El-Ghonemy (2021). To optimize the inoculum size, incubation duration, pH, temperature and substrate concentration, One Factor at a Time (OFAT) was employed. To modify the appropriate moisture level, the fermentation media contained 2 mL of mineral salts solution comprising of 2g potassium dihydrogen phosphate (KH2PO4), 5g ammonium nitrate (NH4NO3), 1g sodium chloride (NaCl) and 1g magnesium dihydrogen sulfate (MgSO4.7H2O) in a liter of distilled water. All the ingredients were combined, autoclaved for 20 minutes at 121°C, then allowed to cool. After that, spore suspensions were placed on a sterile solid substrate and incubated at different temperatures (El-Ghoney 2021).
Temperature, incubation time, pH, inoculum size and substrate concentration are all key factors in enzyme development and have a significant impact on enzyme activity. The optimization experiments were conducted using one factor analysis with three replicates for each determination according to Batista et al. (2021). Fermentation was observed for 7 days at 30oC. After 24, 48, 72, 96, 120, 144, and 168 hours of incubation, samples were taken to determine amylase activity.
The temperature for incubation was set within the range of 30, 35, and 40oC for the selection of the optimum temperature for amylase production, and the amylase test was performed at the end of the optimum incubation period. Using a pH meter, the pH of the fermenter was modified to several levels ranging from (3.0, 4.0, 5.0, 6.0 and 7.0) using two buffer systems (1N NaOH and 1N HCl). The amylase assay was performed at the previously determined optimum time. Each fermentation medium was fed with varying inoculum sizes (1 × 105 and 1 × 107 spores/mL). Amylase activity was assayed at the end of the optimum incubation period.
Different concentrations of the substrates (groundnut shell) at 1 %, 2 %, 3 %, 4 %, and 5 % (w/v) were used in separate fermentation flask. Amylase activity was assayed at the end of the optimum incubation period. The fermented quantity was combined with 50 mL distilled water at the end of fermentation and stirred for 1 hour on a rotary shaker at 150 rpm. After filtering through Whatman filter paper No. 1, it was centrifuged for 15 minutes at 6000 rpm. The cell-free supernatant was extracted as crude enzyme for further investigation (Olakusehin et al. 2021).
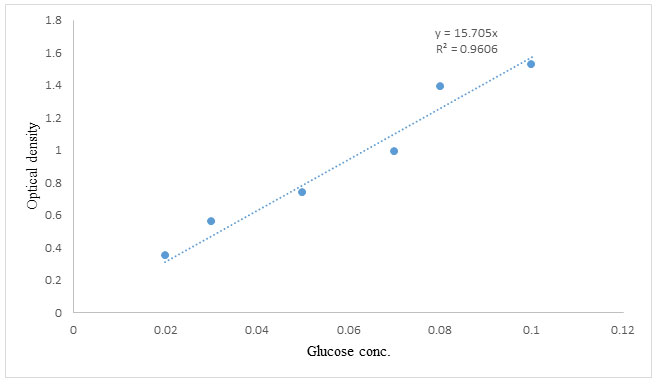
According to Miller’s DNSA assay method, the amount of reducing sugars produced in a mixture containing 1.0 mL soluble starch in phosphate buffer, pH 6.0, and 1.0 mL enzyme extract was measured to determine α-amylase activity (Deshayath et al. 2020). After 10 minutes of incubation in a water bath at 50°C, the reaction was stopped with 1.0 mL of dinitrosalicylic acid (DNSA) reagent and the mixture boiled for 15 minutes. After allowing the test tubes to cool, the absorbance was measured at 540 nm with a UV spectrophotometer. The glucose concentration discharged was compared to a glucose standard. Under assay conditions, one enzyme activity unit (U) was described as the quantity of enzyme that released one mole of reducing sugar per minute per milliliter.
RESULTS AND DISCUSSION
The results of this work aimed at optimizing alpha amylase production from locally isolated Aspergillus sp. using agricultural waste (groundnut shell) as substrate are documented in the subsections below:
Isolation and characterization of fungal isolates: Six fungi were isolated from the soil samples and were given the letters M1, M2, M3, M4, M5, and M6. Table 2 lists the macroscopic and microscopic properties of the isolated fungi, whereas Plates 1-6 depict a microscopic perspective of their vegetative structure.
Table 1: Macroscopic and microscopic characteristics of isolated fungi
| Isolate Code | Colonial Characteristics | Morphological Characteristic Under Microscope | Identity |
| M1 | Filamentous with white hyphae, production of black spores was observed on the plate after 72hrs. The reverse of the plate was brown. | Conidiophores were hyaline, erect, simple, thick-walled, enlarged at the apex, forming globose vesicles containing catenulate conidia with conidial heads | Aspergillus niger |
| M2 | Growth was rapid and filled the plate completely within a few days. Colonies were whitish. Dense and cottony which became greyish-brown with age, due to brownish sporangio-spores and brown black sporangia. Mycelia were interwoven | Well-developed hyphae, branched freely, coenocytic. Brown colored, smooth walled. Non-septate and erect sporangiophores developed from the hyphae | Rhizopus stolonifer |
| M3 | Pin like green growth. | Non-Branched conidiophore with bulb end carried conidia. | Aspergillus flavus |
| M4 | Yellow-orange, ochraceous, or buff colonies with restricted growth. | Non-dense colonies, sporulated, amber-colored, flaky texture, white mycelium with yellow to pale orange or gray gold reverse. Strong presence of light brown sclerotia | Aspergillus ochraceus |
| M5 | Colonies showed slow growth, primarily olivaceous-brown to blackish brown, but also brown, grey or buff, suede-like to floccose, and frequently powdery due to abundant conidia production. | Conidiophores were erect, straight or flexuous, unbranched or branched only in the apical region and elongated with geniculate sympodial elongation in some species. | Cladosporium sp. |
| M6 | Colonies were rapidly growing, white, flat, to cream in dry, color and finely suede-like, with no contrary pigment. | The holoarthric fragmentation of undifferentiated hyphae produces chains of hyaline, one-celled, smooth, sub-globose to cylindrical, slimy arthroconidia (ameroconidia). | Geotrichum candidum |
Screening of isolated fungi for α-amylase activity: Three out of the six isolates exhibited obvious zones of clearance on the amylase agar with the highest (greater than 1 cm) occurring in isolates M1 (1.6 cm) and M4 (1.2 cm) which significantly differ from others, as shown in Table 3. The isolate (M1) with the highest zone of clearance was therefore selected for further studies like molecular investigation to confirm the authentication of its identity as shown in Table 4.
Using locally isolated Aspergillus sp., the current study attempts to optimize α-amylase synthesis from groundnut shell. The findings of this study demonstrated that Aspergillus sp. could produce α-amylase from the substrate (groundnut shell). Six (6) distinct fungal isolates were obtained from various samples (Table 2). The isolates’ morphological and microscopic properties were investigated and reported (Table 2). After that, the isolates were tested for α-amylase activity to see if they could generate the enzyme.
The zone of clearance displayed by the different isolates was evaluated using amylase agar, as described in the screening of amylase producing microorganisms’ section above. During the hydrolysis test, the observed zone of clearance revealed that isolate M1 had the largest diameter of 1.6 cm, which was larger than the other isolates (Table 3). The clearance zones created around the colonies suggest that the fungal isolates can produce extracellular amylase. The findings of this investigation corroborate those of Olakusehin et al. (2021), who claimed that Aspergillus flavus S2-OY has amylolytic characteristics. According to Sahnoun et al. (2015), Aspergillus oryzae S2 has amylolytic characteristics.
Further research was conducted using isolate M1 (Aspergillus sp.). The isolate was used to perform time course fermentation under non-optimized conditions of pH 7.0, 2 % substrate concentration, and 1 ×107 spore/mL inoculum size. The results showed that α-amylase activity peaked at 72.3 U/mL on day 5, with a maximum value of 72.3 U/mL. Using Aspergillus sp., a time course fermentation was used to determine and monitor the trend in α-amylase synthesis from groundnut shell. The recoded enzyme yield is quite similar to that published by (Ahmed et al. 2019), who reported amylase activity of 72.4 U/mL after optimizing with OFAT (Ahmed et al. 2019).
The impact of numerous parameters was explored using OFAT, which was used to optimize a wide variety of factors and values. The impact of several parameters several parameters were investigated including pH, incubation time, temperature, substrate concentration and inoculum size. The best variables for α-amylase synthesis were pH 6, inoculum size of 1 × 107 spores/mL, incubation length of 120 h, substrate concentration of 3 percent (w/v), and temperature of 35oC, as determined by an optimization experiment. In a single fermentation, these optimum conditions were used, and the experiment yielded an optimum enzyme yield of 121.3 U/mL. When compared to non-optimized settings, the enzyme activity obtained after optimization was 40 % higher. This is consistent with the findings of (Ahmed et al. 2019), who found amylase activity of 145.4 U/mL after a Response Surface Methodology optimization trial (RSM). The findings of this study shows that groundnut shell can be used to produce value-added products like α-amylase (El-Ghonemy 2021).
Plate 1: Microscopic view of the vegetative structure of Aspergillus niger (x 40)
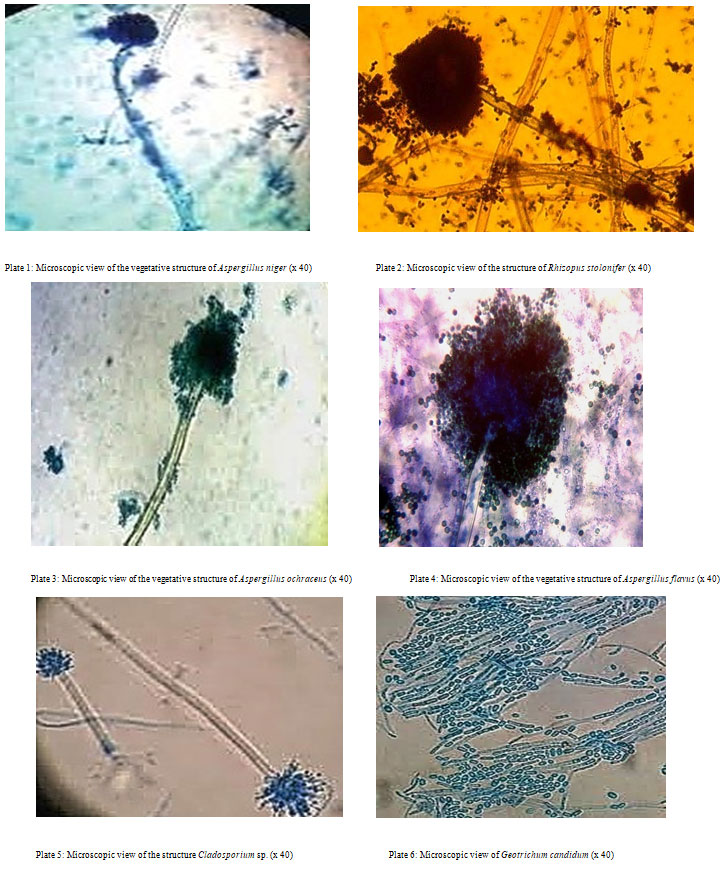
Table 2: Zone of clearance (cm) of different fungal isolates
| Isolate code | Tentative name | Zone of clearance (cm) |
| M1 | Aspergillus niger | 1.6 |
| M2 | Rhizopus stolonifera | 0.5 |
| M3 | Aspergillus ochraceus | 1.0 |
| M4 | Aspergillus flavus | 1.2 |
| M5 | Cladosporium spp. | 0.7 |
| M6 | Geotrichum candidum | – |
Table 3: Molecular confirmation of M1
| Isolate | Organism | Number of Bases | Identity | Accession number | ||
| M1
|
Aspergillus niger
ATCC 16888 |
576 | 100.0 % | NR_111348.1 | ||
α-amylase production by Aspergillus sp.
Time course fermentation: The result of the pre-optimization experiment under the initial conditions of growth (pH 7.0, 2 % substrate concentration and inoculum size of 1 × 107 spores/mL) is shown in Figure 2. It was observed that α-amylase activity picked after 24 h of fermentation reaching its peak (72.3 U/mL) at day 5 before declining (Figure 2).
Effect of optimized environmental condition, substrate, and inoculum size on α-amylase production by Aspergillus sp.
Figure 2: shows the influence of varied incubation periods (days 1-7) on α-amylase production in Aspergillus sp. On day 5, the maximum enzyme activity (72.3 U/mL) was detected.
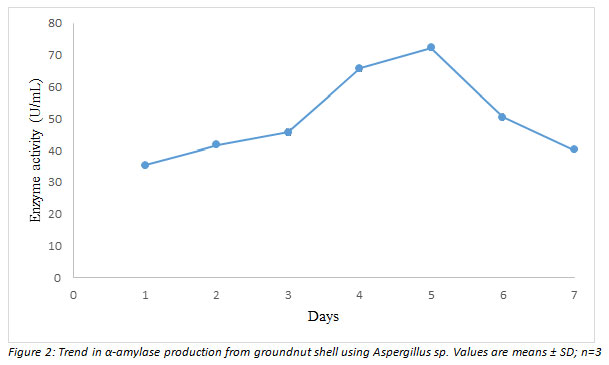
Table 4 shows the effect of temperature (30, 35, and 40 degrees Celsius) on α-amylase production. At 35 degrees Celsius, the greatest enzyme activity was (88.2 U/mL).
Table 5 shows the effect of changing pH (3.0 – 7.0) on α-amylase production. At pH 6, the maximum enzyme production was found (87.3 U/mL).
Table 6 shows the effect of different inoculum sizes (1 × 105 and 1 × 107 spores/mL) on total α-amylase production. Using a 1 × 107 spores/mL inoculum size, the greatest enzyme production was found at (73.9 U/mL).
Table 7 shows the effect of different substrate concentrations (1 % – 5 % w/v) on α-amylase production. At a substrate concentration of 3 % w/v, the greatest enzyme production was obtained (65.1 U/mL).
Table 5. Effect of varying temperature (30, 35, 40°C) on α-amylase production
| Temperature (oC) | Enzyme activity (U/mL) |
| 30 | 65.2 |
| 35 | 88.2 |
| 40 | 71.3 |
Table 6: Effect of varying pH (3.0 – 7.0) on α-amylase production
| (pH) | Enzyme activity (U/mL) |
| 3 | 63.9 |
| 4 | 66.0 |
| 5 | 72.7 |
| 6 | 87.3 |
| 7 | 79.6 |
Table 7: Effect of varying inoculum size (1 × 105 and 1 × 107 spores/mL) on the total α-amylase production
| Inoculum size (spores/mL) | Enzyme activity (U/mL) |
| 1 × 105 | 65.6 |
| 1 × 107 | 73.9 |
Table 8: Effect of varying substrate concentration (1 % – 5 % w/v) on α-amylase production
| Substrate concentration (%) | Enzyme activity (U/mL) |
| 1 | 57.2 |
| 2 | 59.3 |
| 3 | 65.1 |
| 4 | 54.9 |
| 5 | 52.3 |
α-amylase production using optimum conditions
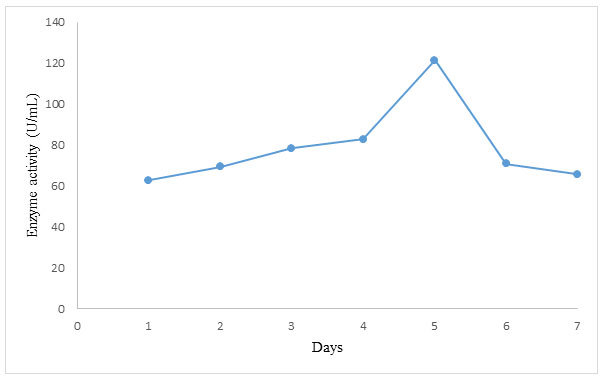
The set of optimum conditions obtained from this study were combined in a single fermentation and the result is shown in Figure 3.
Figure 3: Trend in α-amylase production using set of optimum conditions
Glucose standard curve
CONCLUSION
The findings of the present study have shown that in non-optimized experiments, the maximum α-amylase concentration was 72.3 U/mL, while the optimum concentration was 121.3 U/mL after optimization with the optimum conditions of pH 6, inoculum size of 1×107 spores/mL, incubation period of 120 h, substrate concentration of 3% (w/v), and temperature of 35oC. The use of groundnut shell as an enzyme substrate proved to be highly promising, with reasonable yields. As a result, these findings imply that groundnut shell, which is typically regarded as a waste, might be exploited as a cheap agro-industrial substrate for enzyme production.
REFERENCES
Ahmed, S.A., Abdella, M.A.A., El-Sherbiny, G.M., et al. (2019). Application of one –factor-at-a-time and statistical designs to enhance α-amylase production by a newly isolate Bacillus subtilis strain-MK1. Biocatalysis and Agricultural Biotechnology. 22, 101397.
Almulaiky, Y. Q., Khalil, N. M., El-Shishtawy, R. M., et al. (2021). Hydroxyapatite-decorated ZrO2 for α-amylase immobilization: Toward the enhancement of enzyme stability and reusability. International Journal of Biological Macromolecule, 167:299-308. doi: 10.1016/j.ijbiomac.2020.11.150.
Ani, E., Adekunle, A. A., Kadiri, A. B and Njoku, K. L. (2021). Rhizoremediation of hydrocarbon contaminated soil using Luffa aegyptiaca (Mill) and associated fungi. International journal of Phytoremediation. 23(14):1444-1456. doi: 10.1080/15226514.2021.1901852.
Batista, R. D., Melo, F. G., do Amaral-Santos, C. C. A., de Paula-Elias, F. C., Perna, R. F., Xavier, M. C. A., Villalba Morales, S. A and de Almeida, A. F. (2021). Optimization of β-Fructofuranosidase Production from Agrowaste by Aspergillus carbonarius and Its Application in the Production of Inverted Sugar. Food technology and Biotechnology. 59(3):306-313. doi: 10.17113/ftb.59.03.21.6934.
Chen, L., Chen, W., Zheng, B., et al. (2022). Fermentation of NaHCO3-treated corn germ meal by Bacillus velezensis CL-4 promotes lignocellulose degradation and nutrient utilization. Applied Microbiology and Biotechnology. 17. doi: 10.1007/s00253-022-12130-7.
Deshavath, N. N., Mukherjee, G., Goud, V. V., et al. (2020). Pitfalls in the 3, 5-dinitrosalicylic acid (DNS) assay for the reducing sugars: Interference of furfural and 5-hydroxymethylfurfural. International Journal of Biological Macromolecules. 156:180-185. doi: 10.1016/j.ijbiomac.2020.04.045.
El-Ghonemy, D.H. (2021). Optimization of extracellular ethanol-tolerant β-glucosidase production from a newly isolated Aspergillus sp. DHE7 via solid state fermentation using jojoba meal as substrate: purification and biochemical characterization for biofuel preparation. Journal of Genetic engineering and Biotechnology. 19(1):45. doi: 10.1186/s43141-021-00144-z.
Igbokwe, V. C., Ezugworie, F. N., Onwosi, C. O., et al. (2022). Biochemical biorefinery: A low-cost and non-waste concept for promoting sustainable circular bioeconomy. Journal of Environmental management. 305:114333. doi: 10.1016/j.jenvman.2021.114333.
Kalia, S., Bhattacharya, A., Prajapati, S. K et al. (2021). Utilization of starch effluent from a textile industry as a fungal growth supplement for enhanced α-amylase production for industrial application. Chemosphere. 279:130554. doi: 10.1016/j.chemosphere.2021.130554.
Olakusehin, V.O. and Oyedeji, O. (2021). Production of α-amylase from Aspergillus flavus S2-OY using solid substrate fermentation of potato (Solanum tuberosum L.) peel. International Journal of Biological and Chemical Sciences. 15(5):1950-1967.
Pasin, T. M., Dos Anjos Moreira. E., de Lucas, R. C., et al. (2020). Novel amylase-producing fungus hydrolyzing wheat and brewing residues, Aspergillus carbonarius, discovered in tropical forest remnant. Folia Microbiologica. 65(1):173-184. doi: 10.1007/s12223-019-00720-4.
Sabino, T. P. F., Coelho, N. P. F., Andrade, N. C., et al. (2021). Lignocellulosic materials as soil-cement brick reinforcement. Environmental Science and Pollution Research International, 29(15):21769-21788. doi: 10.1007/s11356-021-17351-3.
Saha, S.P. and Mazumdar, P. (2019). Optimization of process parameter for alpha amylase produced by Bacillus cereus amy3 using one factor at a time (OFAT) and central composite rotatable (CCRD) design-based response surface methodology (RSM). Biocatalysis and Agricultural Biotechnology. 19: 101168.
Sahnoun, M., Kriaa, M., Elgharbi, F., et al. (2015). Aspergillus oryzae S2 alpha-amylase production under solid state fermentation: Optimization of culture conditions. International Journal of Biological Macromolecules. 75: 73-80.
Santana, D. A. R., Scatolino, M. V., Lima, M. D. R., et al. (2021). Pelletizing of lignocellulosic wastes as an environmentally friendly solution for the energy supply: insights on the properties of pellets from Brazilian biomasses. Environmental Science and Pollution Research. 28(9):11598-11617. doi: 10.1007/s11356-020-11401-y.
Shanthala, J., Parveen, G. S and Jambagi, P. K. B. (2022). Genomic-Assisted Breeding for Enhanced Harvestable (Pod) and Consumable (Seed) Product, Yield Productivity in Groundnut (Arachis hypogaea L.). Accelerated Plant Breeding. Volume 4, pp 181-237. doi:10.1007/978-3-030-81107-5-7.
Tela, M., Cresswell, W and Chapman, H. (2021). Pest-removal services provided by birds on subsistence farms in south-eastern Nigeria. PLoS One. 16(8):e0255638.doi: 10.1371/journal.pone.0255638.


