1Division of Biotechnology, Centre for Post-Graduate Studies, Jain University, Bangalore, India
2Division of Biotechnology, Indian Institute of Horticultural Research, Hesaraghatta Lake Post, Hesaraghatta, Bangalore-560089, India
3Division of Floriculture and medicinal Crops, Indian Institute of Horticultural Research, Bangalore-560089, India
Corresponding author Email: aswathiihr@gmail.com
Article Publishing History
Received: 10/07/2018
Accepted After Revision: 19/09/2018
Farmers can harness the full potential of any hybrid only when they get genetically pure seeds of the hybrid. Hence, ensuring the genetic purity of certified seeds of brinjal hybrids is mandatory in India, which is done through field grow out test (GOT) based on the morphological characters of plants grown to maturity. GOT being land and labour intensive, time consuming and influenced by the environment, there is a need to identify rapid and reliable alternatives like DNA based assays. Therefore, the present study was undertaken to identify the SSR markers that could be used to test the hybrid purity of two commercial brinjal hybrids (viz., Arka Anand and Brinjal Asha) and to optimize the minimum sample size that can be used for purity assessment of the brinjal hybrids. Among 120 SSR markers studied, two markers were found to be suitable for testing the purity of these hybrids. The analysis of plant-to-plant variation within the parental lines of all the hybrids, using the identified hybrid specific markers, showed highly homogenous SSR profile, which further indicated the scope of application of these markers in maintenance and purity testing of hybrids and parental lines. These two co-dominant markers can be used as referral markers for unambiguous identification, seed purity testing and protection of the hybrids.
Brinjal, Co-Dominant, Got (Grow Out Test), Hybrid Purity, SSR
Pattanaik A, Reddy D.C. L, Singh T. H, Pandiyaraj P, Chennareddy A. Evaluation of Industrial Effluent and Domestic Sewage Genotoxicity Using Allium Cepa Bioassay. Biosc.Biotech.Res.Comm. 2018;11(3).
Pattanaik A, Reddy D.C. L, Singh T. H, Pandiyaraj P, Chennareddy A. Evaluation of Industrial Effluent and Domestic Sewage Genotoxicity Using Allium Cepa Bioassay. Biosc.Biotech.Res.Comm. 2018;11(3). Available from: https://bit.ly/2lRX50U
Introduction
Brinjal (Solanum melongena L.) identified as eudicot, warm weather crop majorly grows in tropical and subtropical regions of the world and is also known by name eggplant (Schippers 2000; Daunay and Hazra, 2012). Brinjal remains fifth most valued crop of Solanaceous with commercial value of US $10 billion a year after potato, tomato, pepper and tobacco (FAO, 2014). India is the major Brinjal producer with annual production reaching 13.55 million tonnes with a 19.1 mt/ha productivity (NHB database, 2014). Brinjal also remains rich in nutritional contents such as vitamins, minerals and bioactive materials which supply number of benefits to human beings, (Raigon et al., 2008; Plazas et al., 2014b; Docimo et al., 2016).
Although eggplant remains the promising crop for productivity it remains susceptible to number of plant pathogens such as bacterial wilt, fusarium wilt and many others (Rotino et al., 1997). It is also being used as nutrient source by number of pests such as shoot borer, whiteflies, mites, aphids and others (Medakker and Vijayaraghavan, 2007; Rotino et al., 1997). In an attempt related to crop improvement, number of breeding programmes are involving new characters to create promising varieties as F1 generation imparting better shelf life, disease resistance and nutritional qualities (Daunay and Hazra, 2012). In many seed industries, Hybrid seed production of the Brinjal is carried out by hand emasculation and pollination; still brinjal undergoes cross-pollination to change the genetic purity by involving foreign pollens.
The traditional Grow Out Test (GOT) is based on the genetically induced morphological and biochemical features expressed at suitable stages of development. Grow out test (GOT) is an important method for genotypic identification through adult plant phenotype (Arus, 1983). The physical features are studied in detail and all the other plants i.e. off-types, pollen shredder, objectionable and diseased plants, etc are traced out. GOT has got certain limitations, as it is an expensive procedure. GOTs take up a full growing season, which often results in late entry of seed in market. In addition to GOT, number of biochemical markers are used such as isozymes and seed storage proteins, but they also remained restricted in their usage (Dadlani et al., 1997; Mehetre and Dahat, 2001; Borle et al., 2007 and Rakshit et al., 2008). Keeping in view, scientific community is now moving towards DNA markers which provides diversity in approach with the type of methodology adopted such as simple sequence repeats (Rana, 2003; Dongre and Parkhi, 2005; Saravanan et al., 2007), Amplified fragment length polymorphism (Rana and Bhat, 2004), Random amplified polymorphic DNA (Geng et al., 1995; Venu, 2001, Rao et al., 2002). Restriction fragment length polymorphism (Pendse et al., 2001; Dongre and Parkhi, 2005) and ISSR marker (Dongre and Parkhi, 2005; Rana, 2006). These are now widely used in hybrid purity check program.
Among all DNA markers, now SSR markers are found to be promising with its co-dominant, polymorphic, discriminative, reliable and repeatable features and can be standardized for Distinctness, Uniformity and Stability (DUS) testing (UPOV, 1997). This marker (SSR) undoubtedly overcome the problem of errors in morphological based selections of hybrids which is occurring due to plant growth stage and environmental fluctuation.Even though molecular marker plays such an important role in identifying the hybrid purity, there are no guidelines available on the use of specific number of markers & sample size for 90, 95, & 99% purity in comparison with the GOT (Grow-Out Test), where 400 seeds were used for 95% purity. There is an urgent need for this method to be standardized for large scale “Hybrid Purity Test”. Hence, the present study was undertaken with the objective of optimizing the minimum sample size that can be used for purity assessment of the brinjal hybrids. The results obtained using SSR markers were then compared with those from a GOT performed on the various sample size and the percentage of genetic purity was calculated for both GOT and SSR analysis.
Materials And Methods
Plant Materials
The present study was carried out at Indian Institute of Horticultural Research, Bangalore, India during 2014-2016. Two commercial F1 hybrid brinjal cultivars were tested. Cultivar ‘Arka Anand’ and its parental (female and male) lines were developed in Indian Institute of Horticultural Research, Bangalore, India while ‘Brinjal Asha’ and its related parental (female and male) lines were provided by Noble Seeds Pvt. Ltd., Bangalore, India.
Grow Out Test (GOT)
To validate the conformity of the molecular marker-based estimates of selfed or off-type or outcrossed plants with the actual field morphological data, the experiment was conducted by pooling 95% F1 hybrids mixed with 5% off-types / admixture individually in the following sets of 400, 300, 200 and 100. The parental and four set of seed lots from each hybrid were sown in the greenhouse with a day temperature of 24 + 3 ºC and a night temperature of 18 + 3 ºC in the Indian Institute of Horticultural Research, Bangalore. One-month-old seedlings were transplanted to the polyhouse in 5m row with 10cm plant to plant and 45cm row to row spacing. Regular irrigation, fertilization, staking and crop protection measures were adopted, and purity visual evaluation was conducted based on the important morphological characters throughout the growth period. The details on morphological traits that have been recorded to distinguish the true hybrids from off types for all the two hybrids along with their parents are furnished in table 1. The genetic purity of hybrids was calculated as:
((Total number of plants-number of off types))
Hybrid purity (%) = ×100
(Total number of plants)
| Table 1: Morphological characters used to identify the selfed/offtypes during the Grow- out Test. | ||||
| Brinjal (Arka Anand) | ||||
| Sl. No. | Morphological characters | Female parent | Male parent | Hybrid (Brinjal Asha) |
| 1. | Plant: Growth habit | Semi erect | Semi erect | Erect |
| 2. | Stem hairiness | Present | Slightly present | Present |
| 3. | Leaf hairiness | Present | Absent | Slightly Present |
| 4. | Leaf colour | Dark green | Light green | Light green |
| 5. | Flower:colour | Dark purple | Dark purple | Light purple |
| 6. | Flower:petal no. | 5 | 6 | 6 |
| 7. | Calyx:spininess | Present | Absent | Absent |
| 8. | Fruit:colour of calyx | Purplish | Greenish | Light Greenish |
| 9. | Fruit: General shape | Obovate | Ovoid | Ovoid |
| 10. | Fruit: Colour | Dark purple | Dark purple | Light purple |
| 11. | Fruit: Stripes | Absent | Present | present |
| Brinjal (Asha) | ||||
| Sl. No. | Morphological characters | Female parent | Male parent | Hybrid |
| 1. | Plant: Growth habit | Semi erect | Semi erect | Erect |
| 2. | Stem hairiness | Present | Slightly present | Present |
| 3. | Leaf hairiness | Present | Absent | Slightly Present |
| 4. | Leaf colour | Dark green | Light green | Light green |
| 5. | Flower:colour | Dark purple | Dark purple | Light purple |
| 6. | Flower:petal no. | 5 | 6 | 6 |
| 7. | Calyx:spininess | Present | Absent | Absent |
| 8. | Fruit:colour of calyx | Purplish | Greenish | Light Greenish |
| 9. | Fruit: General shape | Obovate | Ovoid | Ovoid |
| 10. | Fruit: Colour | Dark purple | Dark purple | Light purple |
| 11. | Fruit: Stripes | Absent | Present | present |
Dna Extraction
Total genomic DNA was extracted from young and fresh leaves using modified CTAB method (Doyle & Doyle, 1990). The quality and quantity of isolated genomic DNA was checked using 0.8% Agarose gel electrophoresis and Gene Quant UV Spectrophotometer (GE Health Care Bio-Sciences Ltd., Bengaluru, India) respectively. After quantification, the final concentration of DNA was adjusted to 20ng/μl and used as template DNA for PCR amplification.
| Table 2: Comparison of hybrid purity assessment based on GOT and SSR analysis with different sample size in Brinjal hybrids. | ||||
| Brinjal (Arka Anand) | ||||
| Sample size | GOT (%) | Putative false hybrid individuals | SSR Analysis (%) | Putative false hybrid individuals |
| 400 | 95 | 20 | 95 | 20 |
| 300 | 95 | 15 | 94.7 | 16 |
| 200 | 95 | 10 | 95 | 10 |
| 100 | 95 | 5 | 95 | 5 |
| Brinjal (Asha) | ||||
| Primer 1 | ||||
| Sample size | GOT(%) | Putative false hybrid individuals | SSR Analysis (%) | Putative false hybrid individuals |
| 400 | 95 | 20 | 95 | 20 |
| 300 | 95 | 15 | 94.3 | 17 |
| 200 | 95 | 10 | 95 | 10 |
| 100 | 95 | 5 | 95 | 5 |
| Primer 2 | ||||
| Sample size | GOT(%) | Putative false hybrid individuals | SSR Analysis (%) | Putative false hybrid individuals |
| 400 | 95 | 20 | 95 | 20 |
| 300 | 95 | 15 | 94.3 | 17 |
| 200 | 95 | 10 | 95 | 10 |
| 100 | 95 | 5 | 95 | 5 |
Pcr Amplification Profile
A total of 120 SSR markers were screened for the detection of polymorphism within the parents of Arka Anand and Brinjal Asha. Later the polymorphic SSR markers were tested on F1 population to determine the discriminatory and stability of F1 plants. The PCR assay was performed in a 25μl volume containing 2.5μl of 10X PCR buffer, 2.5 µL of 20mM MgCl2, 2.5µL of 1mM dNTP mix, 2.5µL of 5pmol of each (F&R) primer, 2.5µl of 20ng/µl of template DNA, 0.3µl of 3 units of Taq DNA polymerase and 9.67µL of double distilled water to a total volume of 25µL. The amplification was carried out in a thermocycler (model TC-5000; Bibby Scientific (Asia Limited, Hong Kong). A touch down PCR protocol was optimised in brinjal with a temperature program consisting of the initial denaturation at 95 °C for 30 minutes followed by 10 cycles with a decrease of 1 ºC per cycle of denaturation at 94 °C for 30 seconds, annealing at 60-55 °C for 30 seconds and polymerization at 72 °C for 1 minute followed by 30 cycles of denaturation at 94 °C for 30 seconds, annealing at 55 °C for 30 seconds and polymerization at 72 °C for 1 minute. Final elongation was at 72 °C for 5 min.
The amplification products were analyzed by electrophoresis in a 3% (w/v) agarose gel containing 0.5 µg/ml ethidium bromide in 45 mM Tris-borate- (1mM) EDTA buffer, pH 8.0. 25 µl of PCR products was loaded into the well after adding 3µl of loading dye (50% (w/v) glycerol and 50% (w/v) BPB). The 100bp DNA ladder (3B bioscience, Spain) was used to calculate PCR product size. Electrophoresis was carried out at 70V for 1hr. The amplified fragments were visualized with UV transilluminator (Syngene, USA) and documented using UV-Pro gel documentation system.
Results and Discussion
In the Grow-out Test, purity evaluation was conducted based on morphological traits. In the present study, plants from four different sample sizes (400, 300, 200 and 100) of ‘Arka Anand and Brinjal Asha’ were studied individually to determine if they were true-to-type for ten morphological characters in the GOT (Table 1). In case of Arka Anand, out of the ten morphological characters analysed; Flower petal no., leaf colour, leaf length, fruit shape and fruit colour exhibited the maximum variation (Figure 1). The percentage of hybrid purity in GOT assay for 400, 300, 200 and 100 sample sizes of Arka Anand was calculated to be 95% (Table 2). In case of Brinjal Asha, out of the ten morphological characters analysed; flower colour, flower petal no., calyx spininess, fruit shape, colour of calyx, fruit colour and presence of fruit stripes exhibited the maximum variation (Figure 2). The characters of few individuals shown deviation from the standard characters were identified as off-type and they were similar to those of the female parental type. The percentage of hybrid purity in GOT assay for 400, 300, 200 and 100 sample sizes of Brinjal Asha was calculated to be 93%, 95%, 95%, 94% respectively (Table 2).
 |
Figure 1 |
 |
Figure 1a |
 |
Figure 1c |
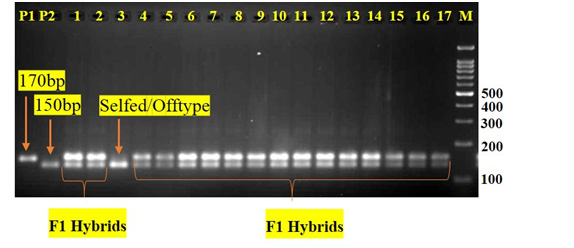
In the PCR-based assay, a total of 120 pairs of SSR markers were tested, with twenty-one markers (Table 3) showing polymorphism between the parental lines. Out of 21 polymorphic markers, two SSR markers (eme08D09 & emb01F16) was found to be Co-dominant. Using eme08D09, we were able to determine hybrid purity of two Brinjal hybrids (Arka Anand & Brinjal Asha) resulted in amplification of 210bp female-specific amplicon (FSA) as well as 230bp male-specific amplicon (MSA). On the other hand, emb01F16 was only useful for determining hybrid purity of one hybrid (Brinjal Asha) resulted in amplification of 150bp female- specific amplicon (FSA) as well as 170bp male-specific amplicon (MSA). Further, these co-dominant markers (eme08D09 & emb01F16) was tested on all the four-sample size of each hybrids to detect the heterozygosity of the hybrids. In 400 hybrid sample size of Arka Anand, the percentage of hybrid purity in SSR analysis was calculated to be 95% (Table 2). Similarly, in the case of 300, 200 and 100 hybrid sample size of Arka Anand, the percentage of hybrid purity was calculated to be 94.7%, 95% and 95%, respectively (Table 2). In 400 hybrid sample size of Brinjal Asha, the percentage of hybrid purity in SSR analysis was calculated to be 95% (Table 2). Similarly, in the case of 300, 200 and 100 hybrid sample size of Brinjal Asha, the percentage of hybrid purity was calculated to be 94.3%, 95% and 95%, respectively (Table 2).
 |
Figure 2a: Variability in morphological parameters of Brinjal hybrid Asha and its parents. |
 |
Figure 2b |
 |
Figure 2c |
 |
Figure 2d |
Based on the results obtained from present study, it is clear that there is a need to critically assess the hybrid purity of popular cultivars of premium quality at each and every stage of seed multiplication and processing with the help of molecular markers so that, seeds cultivated by farmers are true-to-type and fetches premium price. In the present study where, simultaneous laboratory and field study has been done for hybrid purity check of brinjal hybrids, that to in the past number of similar approaches gain promising results and eliminated any bias factor in laboratory analysis (Smith and Wych, 1986; Orman et al., 1991). As Singh and Singhal (1999); Tunwar and Singh (1988) recommended to use minimum 400 individual seeds for purity testing as per Indian minimum seed certification standards, we have observed that by using SSR primers (eme08D09 and/or emb01F16) with only 100 seeds sample size; hybrid purity could be calculated upto 95% percentage which was in agreement with the results obtained via, 400 sample sizes tested in GOT. Hence results indicated that instead of handling 400 samples in GOT, use of only 100 seeds in SSR analysis proving useful with less time and cost concerned approach.
| Table 3: List of polymorphic SSR markers used in the study | ||
| Sl. no. | Primer Name | Primer Sequence 5’ – 3’ |
| 1 | emi04H10 F | ATCGGAGCAAGAGACATTAGATGC |
| R | GTTTCCAACACAGTCCCCAATACAACAA | |
| 2 | emb01D10 F | AAGAATCGGTCCTCTTTGCATTGT |
| R | TGCTTTTCACCTCTCCGCTATCTC | |
| 3 | eme08D09 F | ATGGATTAGCATGTGGAGGACTGAA |
| R | GTTTCATGGTAGGTGGAGACAGAACCA | |
| 4 | emf21K08 F | ATCAATGACACCCAAAACCCATTT |
| R | GTTTGAAAACCCAATACAAATCCGA | |
| 5 | emg11M21 F | ATAGCCTACTGCCTTCAAGACCAT |
| R | GTTTCCTACGTCCAGTCCCCTTAGGT | |
| 6 | emb01J19 F | GACAGGGATAGGGGTACGGATAGG |
| R | ATCCATGTGATGCCTCGATTTTCT | |
| 7 | emb01F16 F | AAAACAGAAGCAAAGTCGGCAGTC |
| R | GTCCACCAACACCTTACCATCCTC | |
| 8 | emi06A04 F | ATTTGGGACAAATGTGGGTGAGAC |
| R | GTTTCCACGCTACTTAGGGGACTCAA | |
| 9 | eme03F04 F | ATATGACGACAGACGTAAAGCGACC |
| R | GTTTCAGAGTTTTGCCATCTGTGTCGAG | |
| 10 | emf11B07a F | ACGAGAGTTGCTACAGTTAAGGGG |
| R | GTTTGGGGACCAAAGTGTATTTTCAAGG | |
| 11 | emk04H07 F | ATTTGGCTGGGTTGTTGGTCTAGT |
| R | GTTTGGCCCAATTACTCAAATACCCTG | |
| 12 | emf21C11 F | AGGTTGGAGCCATGATTACTTGAA |
| R | GTTTGCTACCTATCAAACAGGCGGAA | |
| 13 | emf01O04 F | ATCCGTTGATACTAGCCGTTGCCT |
| R | GTTTCACCCGGTATGAGTGTATCCC | |
| 14 | emk04N11 F | ATCTCCCCCTCAACTTTGAACAAT |
| R | GTTTGTGTGATATAGCCCAACAATTCAC | |
| 15 | emh11N11 F | ATTCAGTTCTTCGCTTTGGAGCTT |
| R | GTTTCCAAACCCGACCCATCCTAAATAA | |
| 16 | eme05G05 F | ACAAGAAAGAGGAGCTGGGGAAATTG |
| R | GTTTCCTTCTTGGGAAGACAACTTATCA | |
| 17 | eme01D03 F | ACAAGAATCGGTCCTCTTTGCATTGT |
| R | GTTTGCTTTTCACCTCTCCGCTATCTC | |
| 18 | emg11P03 F | ACTCGCCTCTCTCAATCTTTCTTG |
| R | GTTTCAATATAACCTCGGCTATGAGACCC | |
| 19 | emd01C04 F | ACCTGCATGAAATGTGTTTGAGTG |
| R | GTTTGGGTCTTTTCATCTCAAAATGGG | |
| 20 | emh05H12 F | AGTCACTGCTCTTAGTTTCTGCAA |
| R | GTTTCAGAGCAGCGATCCTTTCTTCATT | |
| 21 | emd01A01 F | ACAGCAAAATGCCTAATGACAGCACA |
| R | GTTTATGCCTGACTCTGCTTGTGCCTA | |
The present study also recorded the same results with SSR markers when tested with sample size of 400, 300 and 200. Hence instead of using 70 days long protocol involved with GOT, by applying few days protocol of SSR with the given primers; it will assure the early releases of hybrid seeds. As per study results of field grow out tests and SSR markers of individual seeds; highly significant correlation does exist between two studies and more reliability to the latter technique. As the results were fairly consistent; hence SSR markers study can be recommended as a supplementary technique for making speedy decision to accept or reject hybrid seed lots based on contamination.
GOT certainly delays the whole process of decision making, packaging and marketing of the commercial seed. Thus, farmers did not get the hybrid seeds at right time for sowing, resultant precluding the immediate cultivation of the hybrid seed produced. In addition to capital invested on hybrid seed production and additional expenditure incurred on storage of hybrid seed, GOT ultimately increases the hybrid seed cost (Nandakumar et al, 2004). Similar to present study, use of SSR markers for genetic purity testing has been demonstrated in rice (Nandakumar et al., 2004); in maize (Wang et al., 2002) and in sunflower (Pallavi et al., 2011) and overall data remained comparable with field grow out test. Similar to SSR marker applied in present study with minimum sample size; number of workers with other plants have applied different sample size of seeds for RAPD primers for example: 400 seeds in chicory (Bellamy et al., 1998), 120 in canola (Marshall et al., 1994), 40 in tomato (Rom et al., 1995), 30 in Chinese cabbage (Meng et al., 1998) and 10-20 in pepper (Ballester and Vicente, 1998) to assess the genetic purity of the hybrid seeds.
Overall data of the present study have showcased that SSR markers are quick, reliable and results are mostly consistent with morphological analysis in the field study. Markers reported in the study could be involved for routine genetic purity testing of Arka Anand and Brinjal Asha hybrids. The SSR markers developed through this study will be useful for seed industry to select appropriate marker combinations and assess genetic purity of the crop.
Acknowledgements
This research was supported by, Indian Institute of Horticulture Research, Bangalore. Thanks to Dr. C. Aswath and Dr. D. C. L Reddy for their continuous guidance and support in the completion of this research.
References
Arus P. (1983). Genetic purity of commercial seed lots. In Isozymes in Plant Genetics and Breeding (Part A), (eds. S.D. Tanksley and T.J. Orton), Elsevier, Amsterdam, Pp 415–423.
Ballester J, and Vicente M.C. (1998). Determination of F1 hybrid seed purity in pepper using PCR-based markers. Euphytica, Vol. 103(2), Pp 223-226.
Bellamy A, Vedel F and Bannerot H. (1996). Varietal identification in Chicorium intybus L. and determination of genetic purity of F1 hybrid seed samples, based on RAPD markers. The Journal of Plant Breeding, Vol. 115, Pp 128-132.
Borle U.M.,. Mehetre S.S. and Harer P.N. (2007). Seed protein banding pattern studies on different cytosteriles, their maintainers, fertility restores and hybrids of upland cotton. Journal of Maharashtra Agricultural University, Vol. 32, Pp 208–
210.
Dadlani M, Vashist V, Singh D.P. and Varrier A. (1997). A comparison of field Grow out test and electrophoresis methods for testing genetic purity of cotton hybrid seed. Seed Research, Vol. 24, Pp 160–167.
Daunay M.C. and Hazra P. (2012). “Eggplant,” in Handbook of Vegetables, eds Peter KV and Hazra P (Houston, TX: Studium Press), 257–322.
Docimo T, Francese G, Ruggiero A, Batelli G, De Palma M, Bassolino L, Toppino L, Rotino G.L., Mennella G, Tucci M. (2016). Phenylpropanoids accumulation in eggplant fruit: characterization of biosynthetic genes and regulation by a MYB transcription factor. Frontiers in Plant Science, Vol. 6, Pp 1233.
Dongre A and Parkhi V. (2005). Identification of cotton hybrid through the combination of PCR based RAPD, ISSR and microsatellite markers. Journal of Plant Biochemistry and Biotechnology, Vol. 14, Pp 53–55.
Doyle JJ and J. L. Doyle. (1990). Isolation of Plant DNA from Fresh Tissue. Focus, Vol. 12(1), Pp 13-15.
FAO. (2014). FAOSTAT Production Databases. Available online at: http://www.faostat.fao.org (Accessed January 30, 2017).
Geng C.D., Gong Z.Z., Huang J.Q. and Dang Z.L. (1995). Identification of differences between cotton cultivars (G. hirsutum) using RAPD method. Jiangsu Journal of Agricultural Science, Vol. 11 Pp 21–24.
Marshall P, Marchand M.C., Lisieczko Z and Landry B.S. (1994). A simple method to estimate the percentage of hybridity in canola (Brassica napus) F 1 hybrids. Theoretical and Applied Genetics, Vol. 89(7-8), Pp 853-858.
Medakker A and Vijayaraghavan V. (2007). Successful commercialization of insect-resistant eggplant by a public–private partnership: reaching and benefiting resource-poor farmers. In: Intellectual Property Management in Health and Agricultural Innovation: A Handbook of Best Practices, Krattiger A, Mahoney RT, Nelsen L, Thomson JA, Bennett AB, Satyanarayana K, Graff GD, Fernandez C, and Kowalski SP (Oxford: MIHR and Davis; PIPRA), Available online at: www.ipHandbook.org.
Mehetre S and Dahat D. (2001). Page of seed proteins as a technique for varietal identification of Gossypium spp.: A review, Journal of Maharashtra Agricultural University, Vol. 26, Pp 233–238.
Meng, J, S. Shi, L. Gan, Z. Li and X. Qu. (1998). The production of yellow seeded Brassica napus (AACC) through crossing inter-specific hybrids of B. campestris (AA) and B. carinata (BBCC), with B. napus. Euphytica, Vol. 103, Pp 329-333.
Nandakumar N, Singh A.K., Sharma R.K., Mohapatra T, Prabhu K.V. and Zaman F.U. (2004). Molecular fingerprinting of hybrids and assessment of genetic purity of hybrid seeds in rice using microsatellite markers. Euphytica, Vol. 136, Pp 257–264
NHB database. (2014). http://nhb.gov.in/statistics/Publication/HorticultureAtaGlance2017fornetuplod(2).pdf
Pallavi H.M., Rame Gowda, Vishwanath K, Shadakshari Y.G. and Bhanuprakash K. (2011). Identification of SSR markers for hybridity and seed genetic purity testing in sunflower (Helianthus annuus L.). Seed Science & Technology, Vol. 39, Pp 259-264.
Pendse R, Malhotra S, Pawar S and Krishna T. (2001). Use of DNA markers for identifying inbreds and hybrid seeds in cotton (Gossypium hirsutum L.). Seed Science & Technology, Vol. 29, Pp 503–508.
Plazas M, Prohens J, Cunat A.N., Vilanova S, Gramazio P, Herraiz F.J. and Andujar I. (2014). Reducing capacity, chlorogenic acid content and biological activity in a collection of scarlet (Solanum aethiopicum) and gboma (S. macrocarpon) eggplants. International Journal of Molecular Sciences, Vol. 15(10), Pp 17221–17241.
Raigon M.D., Prohens J, Munoz-Falcon J.E. and Nuez F. (2008). Comparison of eggplant landraces and commercial varieties for fruit content of phenolics, minerals, dry matter and protein. Journal of Food Composition and Analysis, Vol. 21, Pp 370–376.
Rakshit A, Vashisht V, Rakshit S, Dadlani M. (2008). Electrophoresis technique for varietal identification and genetic purity in hybrid cotton (Gossypium hirsutum L.). Seed Research, Vol. 36, Pp 28–32.
Rana M.K. (2003). C.F. Use of DNA markers for assessing purity of parental lines and hybrids in various crops, IARI, New Delhi.
Rana M.K., Singh S and Bhat K.V. (2006). RAPD, STMS and ISSR markers for genetic diversity and hybrid seed purity testing in cotton. Seed Science & Technology, Vol. 35, Pp 709–721.
Rao R.G.S, Dadlani M and Mahapatra T. (2002). F1 hybrid seed analysis by RAPD markers in cotton hybrid (Gossypium spp.), Seed Technology News, 32, 162.
Rom M, Bar M, Rom A and Polowsky. (1995). Purity control of F1 hybrid tomato cultivars by RAPD markers. Plant Breeding. Vol. 114, Pp 188-190.
Rotino G.L., Perri E, Acciarri N, Sunseri F, Arpaia S. (1997). Development of eggplant varietal resistance to insects and diseases via plant breeding. Advances in Horticultural Science, Vol. 11, Pp 193–201.
Saravanan N, Ram S, Thiruvengadam V, Ravikesavan R and Raveendran T. (2007). Production and fertility restoration of an interspecific Hybrid between Gossypium hirsutum L. and G. raimondii U. CytologIia, Vol. 72, Pp 195–203.
Schippers R.R. (2000). African indigenous vegetables, an overview of the cultivated species, Natural resources Institute, Chatham, UK.
Singh D.P. and Singhal N.C. (1999). Optimum size for grow out test of cotton hybrids. Seed Research, Vol. 27 (2), Pp 135-139.
Smith J.S.C. and Wych R.D. (1986). The identification of female selfs in hybrids of maize using electrophoresis and morphology. Seed Science & Technology, Vol. 14, Pp 1-8.
Tunwar N.S.and Singh S.V. (1988). Indian Minimum Seed Certification Standards published by, The Central Seed Certification Board, Department of Agriculture and Corporation, Ministry of Agriculture, Government of India.
UPOV. (1997). http://www.upov.int/edocs/tgdocs/en/tg080.pdf
Venu A. (2001). Development of molecular marker-based seed identification procedure for cotton parents and hybrids (Gossypium spp.). M. Sc. Thesis, Tamil Nadu Agricultural University, Coimbatore, India.
Wang J, Zhong G.Y., Chin E.C.L., Register J.C., Riley R.D., Nieber W.S. and Smith J.S.C. (2002). Identification of parents of F1 hybrids through SSR profiling of maternal and hybrid tissue. Euphytica, Vol. 124, Pp 29-34.