1Post Graduate and Research Department of Zoology, Nirmala College for Women, Coimbatore, Tamil Nadu, India.
2Department of Zoology, Vivekanandha College of Arts and Sciences, Tiruchengode, Namakkal, Tamil Nadu, India.
Corresponding author email: neethujeni@gmail.com
Article Publishing History
Received: 15/09/2021
Accepted After Revision: 19/12/2021
Biofilms are species rich, partially due to highly effective powers of diffusion of the microorganisms and have wide tolerance to marine environmental conditions. Characteristically, the first organisms to respond to and convalesce from stress. The present study aims to isolate and identify the biofilm forming bacterial species, collected from surface water and substratum of the ship hull for four seasons (at Chinnamuttom fishing harbor, Southeast coast of India, during June 2015 to May 2016. Among the mean concentration of bacterial isolates of both water and substratum of the station, maximum in monsoon and minimum in summer seasons. Totally 16 isolates were obtained, based on the adherence property, 8 isolates from surface water and 8 isolates from substratum of the ship hull.
The samples were plated on Zobell marine agar medium for bacterial isolates of study area. The isolates of Bacillus sp., Flavobacterium sp., Pseudomonas sp., Aeromonas sp., Micrococcus sp., Vibrio sp., Salmonella sp., Staphylococcus sp., Shegella sp., Klebsiella sp., Corynebacterium sp., Enterobacter sp., Chromohalobacter sp., Bacillus sp., Escherichia coli and Bacilus sp. were reported in all the seasons at study area. The major diverse bacterial isolates were further characterized through morphological and molecular identification. Based on 16S rRNA gene sequences. Biofilm bacterial isolates were confirmed as Bacillus sp., and Pseudomonas sp.
Bacterial isolates, Biofilm, Chinnamuttam harbor, Substratum, Water.
Nithya P, Dhanalakshmi B, Vijayalakshmi S. On the Diversity of Biofilm-Forming Bacteria in Chinnamuttom Harbor of Southeast India. Biosc.Biotech.Res.Comm. 2021;14(4).
Nithya P, Dhanalakshmi B, Vijayalakshmi S. On the Diversity of Biofilm-Forming Bacteria in Chinnamuttom Harbor of Southeast India. Biosc.Biotech.Res.Comm. 2021;14(4). Available from: <a href=”https://bit.ly/318sKQX“>https://bit.ly/318sKQX</a>
Copyright © Nithya et al., This is an open access article distributed under the terms of the Creative Commons Attribution License (CC-BY) https://creativecommns.org/licenses/by/4.0/, which permits unrestricted use distribution and reproduction in any medium, provide the original author and source are credited.
INTRODUCTION
In marine environment, the harbor waters provide a large diversity of living biomass (microorganisms) and complex mixture of prokaryotes and eukaryotes, which of the most, yet identified/or characterized (Boschker 2005; Biao et al. 2020; Antunes et al. 2020). In marine environment due to forceful opposition of space and living, all surfaces living or innate are susceptible to fouling which is directly related to the colonization and adherence of microorganisms leading to the formation of biofilm formation and bacterial adhesion (Chandran et al. 2020). According to Donlan (2002), the suspended particle surfaces tend to collect and concentrate nutrients by hydrophobic interaction and leads to bacterial colonization.
These colonization on interactive surface supposed to be one of the microbial survival tactics, for increased nutrients access, toxins and antibiotics resistance, and predation protection (Dang and Lovell 2000; Babich et al. 2021). For these advantageous, in submerged substratum, the bacteria are quickly colonized. Bacterial polysaccharides and proteins forms biofilms, and provides accessory to other unicellular fouling organisms (Dobretsov et al. 2013; Zheng et al. 2021; Georgiades et al. 2021)
The primarily colonizized bacteria move to or transported to a surface and involves in series of steps (Mayer et al. 1999; Guiamet and Gomez 2005). In any submerged fresh surface of marine system are predominantly colonized by bacteria and diatoms, such as Vibrio spp., E. coli, P. aeruginosa, Shewanella oneidensis and Bacillus subtilus to initiate the complex biofouling process. Bacterial colonization transpires by a two-step process beginning with reversible attachment to the substratum followed through irreversible adhesion. Bacterial growth is protected and supported by biofilms through reducing the effects of external hazards and disinfecting the materials in aquatic environment (Paul and Jeffrey 1985; Lee and Newman 2003; Omoike and Chorover 2004; Muhammad et al. 2020).
During the expansion of colonization on surfaces, bacteria overproduce extracellular polymeric substances (EPS) (Jayathilake et al. 2017). These polymers, particularly EPS, frequently designated to construct the biofilm matrix, serving as a multi-functional element for immobilization of cells, adhesion on the colonized surface, protection and enabling spatial planning of various species within the biofilm (Carvalho 2018). In some respects, biofilm formation smears to human beings in the remedy process of wastewater, degradation of recalcitrant and aquaculture. In other respects, biofilm processed on heat exchangers, pipelines, ship surfaces and other industrial strategies causes serious problems and consumes large amounts of time and money to removing it. Also, biofilm formed on entrenched materials has been related to microbial diseases.
Therefore, control of biofilm formation has been an important topic of interest to date (Lindberg et al. 2001; Balcazar et al. 2015; Carvalho 2018; Krsmanovic et al. 2020). The present study aims to isolate, identify and characterize the biofilm forming bacteria from water and substratum of the Chinnamuttom harbor. Further, the predominant biofilm forming bacterial species of all the four seasons were further characterized through morphological and molecular phylogenesis.
MATERIAL AND METHODS
The biofilm bacterial samples were collected from surface water and substratum of the ship hull in Chinnamuttom fishing harbor, Kanyakumari, Tamil Nadu. All the samples were collected using sterile polythene bags and immediately freeze and brought to the laboratory for further investigation.
For the isolation of bacteria, the collected samples were serially diluted and placed over Zobell’s marine agar standard spread plates, to isolate the Biofilm bacteria. Plating was done in duplicate and incubated at 37°C for 24 -48 hrs. Following incubation, the total number of Colony Forming Unit (CFU) was determined and morphologically different bacterial colonies were selected, purified and sub-cultured on nutrient agar slant supplemented with 2% NaCl. All the bacterial isolates were tested for observance property by inoculating them into sterile seawater containing glass cover slip. The cover slips were removed and stained with 0.4% crystal violet to determine the adherence of bacteria (You et al. 2007). Bacterial isolates which form a slimy layer on the cover slip were selected for further characterization (Yang et al. 2021).
For the morphological and biochemical characterization, the isolated biofilm bacteria characterized using various morphological, biochemical and molecular characters. Gram stain, acid fast stain, spore staining and motility tests were performed for preliminary identification of the isolate (Cappuccino and Sherman 1999). Morphological parameters include colony color and shape followed by biochemical tests as: Indole, Methyl Red, Voges Proskauer, Triple Sugar Iron (TSI) and Citrate utilization were analyzed. (The establishment of the genera and species present in the samples were identified according to their characteristics as outlined in Bergey’ s Manual of Determinative Bacteriology (Holt et al. 1994; Macfaddin 2000).
To study the molecular characteristics, the screened biofilm producing bacterial isolates were identified through partial 16s rRNA gene sequences analysis. The genomic DNA was extracted using the DNA Kit according to the manufacturer’s instructions. 16s rRNA gene sequence was amplified with bacterial universal primers 518F (5’- CCAGCAGCCGCGGTAATACG -3’ and the reverse primer 800R (5’- TACCAGGGTATCTAATCC -3’). The purified PCR amplicons were sequenced and compared with the 16S rRNA gene sequences available in the GenBank database using BLASTN tool 2.2.28 algorithm (Zhang et al. 2000; Gwak and Rho 2020).
For the phylogenetic analysis, all the 4-biofilm producing bacterial isolates nucleotide sequences were aligned with the program CLUSTAL W (Narasimman et al. 2021). The aligned complete 16S rRNA sequences were exposed to phylogenetic analysis using the Molecular Evolutionary Genetic Analysis (MEGA) software Version 7.0. The tree was generated with the Neighbor-Joining algorithm and bootstrap for 550 resampling to ensure robustness and reliability of trees constructed. The assembled complete 16S rRNA sequences of eleven distinguishable biofilm producing bacterial isolates were deposited in public sequences repository NCBI Gene Bank using the BANKIT sequence submission tool (Narasimman et al. 2021).
RESULTS AND DISCUSSION
Enumeration of biofilm producing bacteria: The present investigation of biofouling bacterial species for four seasons were isolated from surface water and substratum of Chinnamuttom fishing harbor, Southeast coast of India. The biofilm bacteria were higher in both water and substratum of the ship hull on monsoon as compared to post-monsoon, pre-monsoon and summer. The maximum biofilm bacteria population in water was recorded (31.46 [×104] CFU ml−1) in monsoon followed by (30.4 [×104] CFU ml−1) in post monsoon, (27.2 [×104] CFU ml−1) pre-monsoon and (16.53 [×104] CFU ml−1) summer. Likely, the estimated biofilm bacteria value in substratum was high (38.4 [×104] CFU ml−1) in monsoon followed by (36.4 [×104] CFU ml−1) post monsoon; (31.46 [×104] CFU ml−1) pre-monsoon and (18.46 [×104] CFU ml−1) summer. Among the recorded mean values of biofilm bacterial populations, in both water and substratum of the ship hull shows the higher population in monsoon and lower in 16.53 [×104] summer. The similar mean concentration of maximum bacterial isolates on monsoon and minimum in summer seasons were found (Kim et al. 2016; Antunes et al. 2020).
Identification of biofilm bacteria: In the present study totally 16 bacterial isolates were obtained based on biochemical characterization. The bacteria were identified as Bacilus sp., Flavobacterium sp., Pseudomonas sp., Aeromonas sp., Micrococcus sp., Vibrio sp., Salmonella sp., Staphylococcus sp., Shegella sp., Klebsiella sp., Corynebacterium sp., Enterobacter sp., Chromohalobacter sp., Bacillus sp., Escherichia coli and Bacilus sp. (Table 1). Based on the adherence property, 8 isolates from surface water and 8 isolates from substratum of the ship hull were obtained and amplified in all the season samples of Chinnamuttam fishing harbor. Sillankorva et al. (2008), Kreig and Hoit (1984) and hilal et al. (2004) reported Pseudomonas sp., Vibrio sp., Aerobacter aerogens and Bacillus sp. from the fouling substratum. Among these bacterial isolates, Bacillus sp., were frequently observed (Bhattarai et al. 2006; Bhattarai et al. 2007; Antunes et al. 2020). Among the total 16 identified bacterial species, only four bacterial isolates (Table 2) species were record in all the seasons.
Diverse bacterial isolates in the present study were due to the nutritive water of harbor environment and interaction with substratum. The isolated biofilm bacterial strains in water and substratum samples indicate, that most of the isolated strains identified as gram negative in nature (Samrot et al. 2021). Among these biofouling bacterial isolates, Bacillus sp. is one of the predominant bacterial species followed by Pseudomonas sp. and others. In accordance to the present study, occurrence of bacterial strains, B. subtilis, Staphylococcus sp., Micrococcus sp., Salmonella sp., Vibrio sp. and Pseudomonas sp., were the biofilm bacteria involving in 128 types of biofouling process. Based on the hydrographical conditions the biotic organism abundance and biomass occur varies in marine environment (Lee and Newman 2003; Omoike and Chorover 2004; Udhayakumar 2010; Antunes et al. 2020).
Table 1. Morphological and biochemical identification of biofilm bacteria
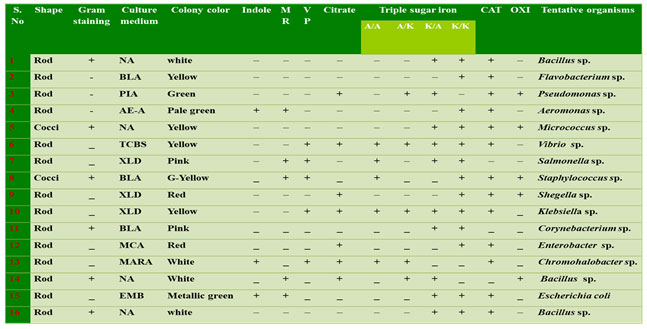
I – Indole ; MR – Methyl red; VP – Voges proskauer; C – Citrate; TSI – Triple sugar Iron; CAT – Catalase; OXI – Oxidase; A/A – Acid slant acidbutt; A/K – Acid slant alkaline butt; K/A – Alkaline slant acid butt; K/K – Alkaline slant alkaline butt. NA – Nutrient Agar; BLA – Blood Agar; AE- A – Aeromonas Agar; TCBS – Thiosulfate citrate Bile Salt Agar; MCA –MacConkey Agar; EMB – Eosin Methylene Blue Agar; XLD – Xylose lysine deoxycholate agar; PIA – Pseudomonas isolation agar.
Motility test: In this present study, the twelve bacterial strains were confirmed as motile forms with a single polar flagellum or multi polar flagellum, remaining four strains was non-motile forms. The motile nature of all the strains was analyzed by commonly used hanging drop method which confirmed that among the 16 identified bacterial strains 4 strains were motile in nature (Figure 1) (Jain et al. 2020).
Bacterial Straining tests: The isolated bacteria were found both cocci and bacilli shaped. Among all the 16 bacterial isolates six gram positive and ten-gram negative bacteria based on absorption of dye in the cell wall of organisms (Table 1). The bacterial strains isolated from water and substratum of the ship hull showed that Gram-positive groups were dominant during all seasons, however, that Gram- negative strains are prevailing on both sites along the harbor environment.
Figure 1: Acid fast stained in dominant bacterial strains (magnification 100X). A- Gram-negative rod-shaped bacteria; B- Gram positive rod-shaped bacteria; C- Gram negative cocci-shaped bacteria; B- Gram positive cocci-shaped bacteria.
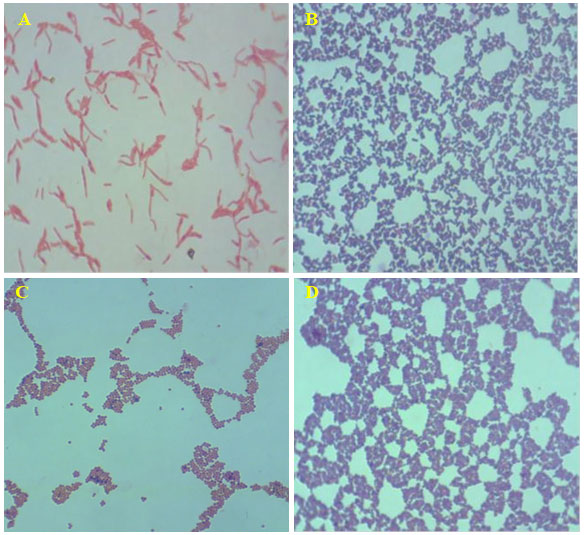
The biochemical test acid fast staining on the 16 isolate showed, bright red to intensive purple, Red, straight or slightly curved rods, happening singly or in small groups, may appear decorated Non-acid fast: blue color (Table 1) and the endospore staining revels the isolates are non-spore form bacteria. The biochemical and straining results revealed that changes in environmental conditions with any season could reflect the capacity of biofilm populations to maintain their diversified functions in an ecosystem. In addition, when observing at the genus/species level within the same phylum, dominances would be expected to change over time.
Although several studies indicate that the presence of biofilms may increase the attachment of macro-foulants, the exact interactive mechanisms are yet to be discovered (Kang et al. 2015). Aeromonas sp. Chromohalobacter sp. Escherichia coli, were given the positive results of indole test. Remaining 13 are the negative strains. The 7 bacterial isolates viz., Pseudomonas sp., Vibrio sp., Shegella sp., Klebsiella sp., Enterobacter sp., Bacillus sp., Chromohalobacter sp. are positive for indole test and only 4 bacterial strains (Aeromonas sp., Salmonella sp., Bacillus sp. and Escherichia coli) showed positive to Methyl red test (Antunes et al. 2020).
Triple sugar test: Table 1 showed the results of Triple Sugar Iron (TSI), from which these strains were isolated, and were alkaline reaction, red colour suggested no fermentation of sucrose or lactose or both and were Acidic reaction, yellow colour suggested to fermentation of lactose or sucrose. Among all isolates of bacterial strains only five bacterial species were positive to Voges proskauer (Vibrio sp., Salmonella sp., Staphylococcus sp., Klebsiella sp. and Chromohalobacter sp.) (Antunes et al. 2020).
Oxidase and Catalase test: The catalase and oxidase rapid test were for preliminary differentiation of isolated bacterial strains for their hydrogen peroxide decomposition and oxygen utilization capacity (Aggarwal et al. 2020). From the oxidase test the presence of cytochrome oxidase enzyme was confirmed in Pseudomonas sp., Micrococcus sp., Staphylococcus sp., Shegella sp. and Bacillus sp. Only three bacterial isolates were negative catalase (Salmonella sp., Bacillus sp. and Corynebacterium sp.). The bacterial isolate SS1 was positive catalase and can ferment glucose, maltose and sucrose and reduced nitrates to nitrites. Rajasekar et al. (2007) reported the presence of Bacillus sp (Rajasekar et al. 2007; Aggarwal et al. 2020).
Molecular identification: The genus confirmed strains were further subjected to 16s rRNA analysis. The sequenced four strains were similar with Bacillus amyloliquefaciens, Bacillus sp., Domibacillus indicus and Psudomonas sp. All the strains were unique to each other in their nucleotide base pairs. Since the strains were isolated from Chinnamuttam harbor environment the strains were named as Bacillus amyloliquefaciens- WS8. Bacillus sp., SS1. Domibacillus indicus – SS8. Pesudomonas sp –WS5 and GenBank accession numbers obtained after submission (Table 2). Based on 16S rRNA gene sequence alignment, the phylogeny tree of biofouling bacterial isolates was assembled with the neighbor-joining method (Dang and Lovell 2000; Babich et al. 2021).
The result from the well-supported phylogeny with high resolution inner branches and bootstraps suggests the existence of bacterial strains from Domibacillusaceae family. The bacteria of Domibacillus species were isolated and dominated in the sediment and water samples of Indian coast. The strains exhibited genetic similarity with genus and species of the Domibacillus indicus. The precise identification of bacterial isolates up to species level is vital since this gives an insight into the bacterial diversity. The identification of biofilm producing, as well as biofilm forming, bacterial isolates in this study was carried out by advanced molecular biology technique based on 16S rRNA gene sequence analysis (Babich et al. 2021).
Table 2. Gene bank accession numbers
| Sample ID | Bacterial isolates | Accession Number |
| WS8 | Bacillus amyloliquefaciens | KX896092 |
| SSI | Bacillus sp. | KX896093 |
| SS8 | Domibacillus indicus | KX896094 |
| WS5 | Psudomonas sp. | KX896091 |
Phylogeny Tree: A phylogenetic evolutionary tree shows the inferred evolutionary relationships among various biological species. The taxa joined in the tree were oblique to inclined from common ancestor. Unrooted trees proved the sprig lumps about ancestry at all. Figure 1 showed the phylogenetic tree of all sample are not similar sequences.
Figure 2: Evolutionary relationships of 11 taxa for 4 different Samples Bacillus amyloliquefaciens, Bacillus sp., Domibacillus indicus and Psudomonass sp., using mega 7.0 version Phylogenetic analysis tool.
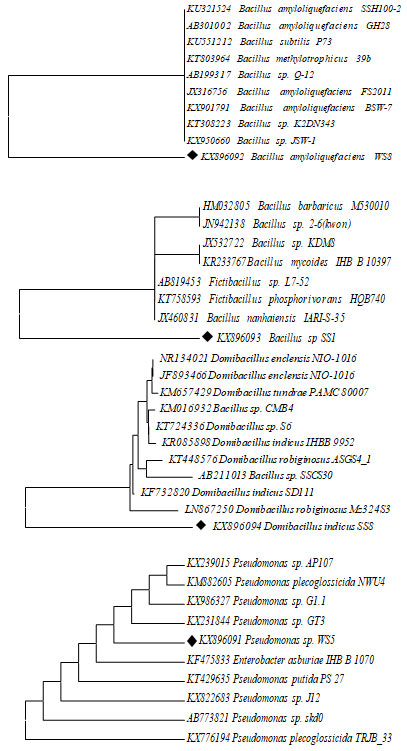
Based on 16S rRNA gene sequences, biofilm bacterial isolates were confirmed as Bacillus sp., and Pseudomonas sp. A molecular technique is one of the important techniques involving ribosomal RNA sequence analysis is commonly used to examine evolutionary interactions within different genera of bacterial isolates. Primary and secondary screening of Streptomyces strains identified by 16s-rDNA gene sequencing and also it has been confirmed that gene sequence data of specific strain with adjoining a parallel score of <97% represents a new species (Reller 2007). The bacterial isolate WS5 was observed as cream-colored circular colonies on Zobell agar medium. The result showed that non motile rod-shaped gram-negative isolate. Various biochemical characteristics of biofilm bacteria were used for their identification.
In the present investigation, it was found that the bacterial isolate was found to be positive results for citrate utilization, catalase and oxidase and negative results for Indole, methyl red, Voges Proskaur and it can acid and gas production form of negative results. It was reported the presence of Pseudomonas sp. Based on amassed whole 16S rRNA gene sequence alignment, the phylogeny tree of these culturable biofilm bacterial isolates was assembled with the neighbor-joining method. This method determines the presence of each bacterial isolate in the phylogeny with bootstrap support. The outcome from high resolution inner twigs recommends the existence of bacterial strains from Pseudomonadaceae family and only one from Enterobacteriaceae (Reller 2007; Babich et al. 2021).
The strains exhibited genetic similarity with genus and species of the Pseudomonas with the cases similarity up to 98% was found. It has been proving that genomic sequence data of separate microbe with a adjacent neighbor displaying a resemblance score of <97% characterizes a new species, the meaning of resemblance scores of >97% is not as clear. The bacterial isolate WS8 and SS8 was perceived as cream color circular colonies on zobell agar medium. The staining report exhibited gram +, rod shaped and motile cells but the strain of SS8 isolates were non-motile. In the present investigation, it was found that the bacterial isolate WS8 was found to be positive for Voges-Proskauer, catalase, oxidase, starch hydrolysis and reduction of nitrate and negative results for Indole production, citrate utilization and hydrolysis of urea. It was reported the presence of Domibacillus indicus (Reller 2007; Babich et al. 2021).
CONCLUSION
The findings of the present study shows that the increased physical parameters in summer may significantly influence to minimize chemical parameters and inorganic nutrients followed by less bacterial count in water and enhance high bacterial load during monsoon season. Hence, the diverse isolates of 16 various biofilm forming bacterial genera were characterized with morphological and biochemical methods. The study found most of biofilm forming bacteria were rod shaped. The seasonal tollerent and dominant species during study period in both water and substratum were Pseudomonas sp., Bacillus amyloliquefaciens, Bacillus sp., and Domibacillus indices, identified and further confirmed through molecular identification. Further investigation of relationship between specific biofilm forming bacterial mass in contrast with macro foulants accumulation and their adherent effect estimation are recommended.
Conflict of Interests: Authors declare no conflicts of interests to disclose.
REFERENCES
Aggarwal D, Kanitkar T, Narouz M et al. (2020). Clinical utility and cost-effectiveness of bacterial 16S rRNA and targeted PCR based diagnostic testing in a UK microbiology laboratory network. Scientific reports, 10(1): 1-9.
Antunes JT, Sousa AG, Azevedo J, et al. (2020). Distinct temporal succession of bacterial communities in early marine biofilms in a Portuguese Atlantic Port. Frontiers in microbiology. 11: 1938.
Babich O, Shevchenko M, Ivanova S, et al. (2021). Antimicrobial Potential of Microorganisms Isolated from the Bottom Sediments of Lake Baikal. Antibiotics. 10(8): 927.
Balcazar JL, Subirats J and Borrego CM (2015). The role of biofilms as environmental reservoirs of antibiotic resistance. Frontiers in microbiology. 6: 1216.
Bhattarai S, Chaudhary RP and Taylor RS (2006). Ethnomedicinal plants used by the people of Manang district, central Nepal. Journal of Ethnobiology and Ethnomedicine. 2(1): 1-8.
Bhattarai S (2007). Antibacterial Activity of Selected Ethnomedicinal Plants of Nawalparasi District, Central Nepal. Research.
Bhosale SH, Nagle VL and Jagtap TG (2002). Antifouling potential of some marine organisms from India against species of Bacillus and Pseudomonas. Marine biotechnology, 4(2), pp.111-118.
Biao Z, Li Y, Xiang KM, et al. (2020). Sediment microbial communities and their potential role as environmental pollution indicators in Xuande Atoll, South China Sea. Frontiers in microbiology. 11:1011.
Cappuccino JG, and Sherman N (2011). Microbiology: A Laboratory Manual, Pearson, (9).
Cappuccino JG and Sherman N (1999). Microbiology, A Laboratory manual, Pearson, (4), 179-182.
Chandran H, Meena M and Sharma K (2020). Microbial biodiversity and bioremediation assessment through omics approaches. Frontiers in Environmental Chemistry. (1): 9. 1:570326
Costerton JW, Cheng KJ, Geesey GG et al. (1987). Bacterial biofilms in nature and disease. Annual Reviews in Microbiology. 41(1): 435-464.
Dang H and Lovell CR (2000). Bacterial primary colonization and early succession on surfaces in marine waters as determined by amplified rRNA gene restriction analysis and sequence analysis of 16S rRNA genes. Applied and environmental microbiology. 66(2): 467-475.
Carvalho CCD (2018). Marine biofilms: a successful microbial strategy with economic implications. Frontiers in marine science. 5:126.
Dobretsov S, Abed RM, Voolstra CR et al. (2013). The effect of surface colour on the formation of marine micro and macrofouling communities. Biofouling. 29(6): 617-627.
Donlan RM (2002). Biofilms: microbial life on surfaces. Emerging infectious diseases. 8(9): 881.
Flemming HC (1999). The role of intermolecular interactions: studies on model systems for bacterial biofilms. International journal of biological macromolecules. 26(1): 3-16.
Georgiades E, Scianni C, Davidson I et al. (2021). The Role of Vessel Biofouling in the Translocation of Marine Pathogens: Management Considerations and Challenges. Frontiers in Marine Science. 8:660125.
Guiamet PS and Gómez DSSG (2005). Laboratory studies of biocorrosion control using traditional and environmentally friendly biocides: an overview. Latin American applied research. 35(4): 295-300.
Gwak HJ, and Rho M (2020). Data-Driven Modeling for Species-Level Taxonomic Assignment From 16S rRNA: Application to Human Microbiomes. Frontiers in microbiology. 11: 2866.
Hilal N, Kochkodan V, Al-Khatib L et al. (2004). Surface modified polymeric membranes to reduce (bio) fouling: a microbiological study using E. coli. Desalination. 167: 293-300.
Holt JG, Krieg NR, Sneath PH et al. (1994). Bergey’s manual of determinative bacteriology, Baltimor: William & Wilkins, (9).
Jain A, Jain R and Jain S (2020). Motility Testing – Hanging Drop Method and Stab. In: Basic Techniques in Biochemistry, Microbiology and Molecular Biology. Springer Protocols Handbooks. Humana, New York, NY.
Jayathilake PG, Jana S, Rushton S et al. (2017). Extracellular polymeric substance production and aggregated bacteria colonization influence the competition of microbes in biofilms. Frontiers in microbiology. 8: 1865.
Kang J, Kang S and Kim Y (2015). Study on the control of marine biofouling developed on the surface of porous ceramics. Journal of the Korean Crystal Growth and Crystal Technology. 25(5): 218-224.
Kim HJ, Jung SW, Lim DI et al. (2016). Effects of temperature and nutrients on changes in genetic diversity of bacterioplankton communities in a semi-closed bay, South Korea. Marine pollution bulletin. 106(1-2): 139-148.
Kreig NR and Holt JG (1984). Bergey’s manual of Systemic Bacteriology. The Archaea and the Deeply branching and phototrophic bacteria, Williams & Wilkins, 1.
Krsmanovic M, Biswas D, Ali H, et al. (2020). Hydrodynamics and surface properties influence biofilm proliferation. Advances in Colloid and Interface Science, 102336.
Lee AK and Newman DK (2003). Microbial iron respiration: impacts on corrosion processes. Applied microbiology and biotechnology. 62(2): 134-139.
Lindberg L, Holmbom B, Vaisanen O et al. (2001). Sugar composition of biofilms produced by paper mill bacteria. Applied microbiology and biotechnology. 55(5): 638-643.
MacFaddin JF (2000). Biochemical tests for identification of medical bacteria.
Mayer C, Moritz R, Kirschner C et al. (2020). Beyond risk: bacterial biofilms and their regulating approaches. Frontiers in microbiology. 11: 28.
Narasimman V, Aavula T, Venkatesan M et al. (2021). Diversity and Molecular Characterization of Marine Halophilic Bacteria from Southeast Coast of India. Geomicrobiology Journal. 38(6): 524-531.
Nichols DS, Sanderson K, Buia A et al. (2002). Bioprospecting and biotechnology in Antarctica. The Antarctic: past, present and future, Antarctic CRC research report. 28: 85-103.
Omoike A and Chorover J (2004). Spectroscopic study of extracellular polymeric substances from Bacillus subtilis: aqueous chemistry and adsorption effects. Biomacromolecules. 5(4): 1219-1230.
Muraleedharan P, Khoda V, Sven G et al. (1999). Incidence of hereditary Citrullinemia and bovine Ieucocyte adhesion deficiency Syndrome in Indian dairy cattle (BOS TAURUS, BOS INDICUS) and buffalo (BUBALUS BUBALIS) Population. Archives Animal Breeding. 42(4): 347-352.
Paul J H and Jeffrey WH (1985). Evidence for separate adhesion mechanisms for hydrophilic and hydrophobic surfaces in Vibrio proteolytica. Applied and Environmental Microbiology. 50(2): 431-437.
Rajasekar A, Babu TG, Pandian SK et al. (2007). Biodegradation and corrosion behavior of manganese oxidizer Bacillus cereus ACE4 in diesel transporting pipeline. Corrosion Science. 49(6): 2694-2710.
Reller LB, Weinstein MP and Petti CA (2007). Detection and identification of microorganisms by gene amplification and sequencing. Clinical infectious diseases. 44(8):1108-1114.
Samrot AV, Abubakar MA, Faradjeva E et al. (2021). Mechanisms and Impact of Biofilms and Targeting of Biofilms Using Bioactive Compounds—A Review. Medicina. 57(8): 839.
Sillankorva S, Neubauer P and Azeredo J (2008). Pseudomonas fluorescens biofilms subjected to phage phiIBB-PF7A. Bmc Biotechnology, 8(1): 1-12.
Swami BS and Udhayakumar M (2010). Seasonal influence on settlement, distribution and diversity of fouling organisms at Mumbai harbor, 39 (1).
Wiencek KM and Fletcher M (1995). Bacterial adhesion to hydroxyl-and methyl-terminated alkanethiol self-assembled monolayers. Journal of bacteriology. 177(8): 1959-1966.
Yang J, Barrila J, Mark OC et al. (2021). Longitudinal characterization of multispecies microbial populations recovered from spaceflight potable water. NPJ biofilms and microbiomes. 7(1), 1-12.
You J, Xue L, Cao X et al. (2007). Inhibition of Vibrio biofilm formation by a marine actinomycete strain A66, Appl. Microbiol.Biotechnol.76: 1137-1144.
Zhang Z, Schwartz S, Wagner L et al. (2000). A greedy algorithm for aligning DNA sequences. Journal of Computational biology. 1: 7(1-2):203-14.


