1Department of Microbiology, School of Science, RK University, Rajkot, India.
2Department of Microbiology, Government Science College, Gandhinagar, India.
Corresponding author email: jignashathumar@gmail.com
Article Publishing History
Received: 18/07/2021
Accepted After Revision: 26/09/2021
Actinomycetes are well known sources of antibiotics, however; recently the focus of antimicrobial research has been turning towards actinomycetes of extreme environments. Therefore, present work would highlight the isolation, identification and characterization of antimicrobial metabolites produced by marine haloalkaliphilic actinomycetes. Saline soil sample was collected from Ghogha coast (Gulf of Khambhat), Bhavnagar, Western India. Isolation was carried out using selective media while identification was done based on morphological, cultural and molecular characterization. The antimicrobial potential was checked by spot inoculation method. Optimization was carried out by the one variable at a time (OVAT) method. The antimicrobial compounds were extracted using ethyl acetate and characterized by GC-MS.
The haloalkaliphilic actinomycetes Nocardiopsis sp. GhM-HA-6 was isolated from saline soil of Ghogha coast using starch agar with 10% w/v NaCl and pH 9 and was identified as Nocardiopsis sp. based on morphology, cultural characteristics and 16S rRNA phylogenetic analysis (NCBI Genbank Accession number: KF384492). The organism showed antimicrobial activity against five Gram positive and three Gram negative bacteria while the isolate didn’t show any antifungal activity. Results of optimization showed that the highest production of antimicrobial compounds was obtained using starch broth with 0.5% w/v starch, 1% w/v yeast extract, 10% w/v NaCl and pH 9. GC-MS analysis of ethyl acetate extract of the isolate showed presence of a total 18 compounds including various antimicrobial compounds like 2, 4-bis (1, 1-dimethylethyl)-Phenol, various types of alkanes and their derivatives. Haloalkaliphilic actinomycete Nocardiopsis sp. GhM-HA-6, from a rarely explored marine habitat, can be a source of antimicrobial compounds with the novel biotechnological applications.
Antimicrobial Activity; Haloalkaliphilic; Gas Chromatography-Mass Spectroscopy;
Marine Actinomycetes; 2, 4-Bis (1, 1-Dimethylethyl)-Phenol
Trivedi N. D, Thumar J. T. On the Bioactivity, Chemical Profiling and 16S rRNA-based phylogeny of Nocardiopsis species isolated from the Gulf of Khambhat, Gujarat, India. Biosc.Biotech.Res.Comm. 2021;14(3).
Trivedi N.D, Thumar J.T. On the Bioactivity, Chemical Profiling and 16S rRNA-based phylogeny of Nocardiopsis species isolated from the Gulf of Khambhat, Gujarat, India. from Hemidesmus indicus. Biosc.Biotech.Res.Comm. 2021;14(3). Available from: <a href=”https://bit.ly/3xTSzOK”>https://bit.ly/3xTSzOK</a>
Copyright © Trivedi and Thumar This is an open access article distributed under the terms of the Creative Commons Attribution License (CC-BY) https://creativecommns.org/licenses/by/4.0/, which permits unrestricted use distribution and reproduction in any medium, provide the original author and source are credited.
INTRODUCTION
Actinobacteria are well known for their ability to produce valuable secondary metabolites such as antibiotics and antimicrobial substances. Amongst genera of actinobacteria, Nocardiopsis genus was primarily described by Meyer in 1976 and was placed in the class of Actinobacteria; subclass Actinobacteridae; order Actinomycetales and family Nocardiopsaceae (Ibrahim et al. 2018). Rainey et al. (1996) defined a new family referred as Nocardiopsaceae considering phylogenetic position, morphological features, and chemotaxonomic properties of Nocardiopsis sp.
This genus includes Gram positive, aerobes, non-acid fast with catalase positive properties (Cook and Meyers 2003; Bennur et al. 2015; Ibrahim et al. 2018). Studies by Kroppenstedt and Evtushenko (2006) showed various features of genus Nocardiopsis which includes the presence of meso-2, 6-diaminopimelic acid, but lack of diagnostically important carbohydrates in cell wall structure, absence of madurose or nocardomycolic acids in whole cell hydrolysates and high GC content in their genomes (Dhakal et al. 2017; Subramani and Sipkema 2019).
Actinobacteria are widely found in soil and physiologically extreme environments such as desert, saline, hyper saline, and alkaline origins. Particularly, Nocardiopsis species are often found from hyper saline environments such as soda lakes and salterns along with Streptomyces species (Mwirichia et al. 2010; Jose and Jebakumar 2012; Kamjam et al. 2017). It is believed that Nocardiopsis genus might be playing a mediating role in breakdown of naturally occurring complex polymers, these species mainly produce extremozymes, compatible solutes, and surfactants (Bennur et al. 2015).
Genus Nocardiopsis has been reported to possess gene clusters for polyketide synthases (PKSs) and non-ribosomal peptide synthetases (NRPSs) and additionally many of the natural isolates have also demonstrated capability antibiotic production (Zitouni et al. 2005; Meklat et al. 2011; Becerril-Espinosa et al. 2013; Romanenko et al. 2013; Schorn et al. 2016; Subramani and Sipkema 2019).
Nocardiopsis sp. often secretes important antimicrobials including polyketides, phenzines, quinoline alkaloids, terphenyls, proteins, thiopeptides, and amines (Bennur et al. 2015). Marine isolates of Nocardiopsis sp. are often reported to produce cyclic hexapeptides which render antimicrobial activity (Wu et al. 2013). Dolak et al. (1980) found the production of a novel antibiotic 3-trehalosamine was reported from soil-derived N. trehalosei. The study also reported production of dopsisamine and thiopeptide antibiotics by N. mutabilis and Nocardiopsis sp. TFS65-07, respectively (Takahashi et al. 1986; Engelhardt et al. 2010a).
Among the nine maritime states in the Indian peninsula, only a few states have been extensively covered for the study of marine actinobacteria for antagonistic properties against different pathogens (Sivakumar et al. 2007). The Gulf of Khambhat is a coastal region situated in the Bhavnagar District of the Indian State Gujarat, which covers approximately 3120 km2 area of mudflat with some rocky (sandstones) intertidal area and a volume of 62,400 million m3 (Ramnathan et al. 2002; Subramani and Sipkema 2019).
Alang and Ghogha coasts of Gulf of Khambhat, India is relatively less explored with respect to antagonistic properties of actinomycetes. Ghogha is a small town situated at the mid-western shore of the Gulf of Khambhat, covering about 4 km long area (21°40’32” to 21°41’18” N and 72°17’5” to 72°16’48 E) in Bhavnagar, Gujarat with unique characteristics of having sandy supratidal zone, rocky-muddy middle intertidal zone and highly muddy lower intertidal zone (Solanki et al. 2016; Subramani and Sipkema 2019).
The uniqueness of this region is its salinity and alkalinity, which harbors various unidentified, unique haloalkaliphilic bacterial species that can potentially produce secondary metabolites. This highlights the urgent need for exploration of untapped locations for novel antimicrobial antagonist producers. In the light of this knowledge, the present study was intended to isolate, screen and characterize the antimicrobial metabolite producing haloalkaliphilic actinomycetes from the Gulf of Khambhat.
MATERIAL AND METHODS
The haloalkaliphilic actinomycete, Nocardiopsis sp. GhM-HA-6 was isolated from saline soil of Ghogha coasts (Gulf of Khambhat), Bhavnagar (Longitude: 72° 11′ E and latitude: 21° 46′ N), India and treated with calcium carbonate and Ringer’s solution. Approximately 10-15g of wet soil sample was mixed with a pinch of calcium carbonate and was dried at 75°C overnight. Next day, 1g of soil sample was added to 10ml of Ringer’s solution. From the supernatant, 0.1 ml of the sample was spread separately on starch agar (Hi-Media, India), containing 10% w/v NaCl and pH 9. The plates were incubated at 30°C; on the sixth day a typical chalky white colony was appeared, the colony characteristics were noted and Gram’s staining was performed.
On the seventh day, colony was picked up and re-streaked on starch agar slants supplemented with 10% w/v NaCl and pH 9 to ensure the purity of the colony (Chakrabarti 1998). The culture was maintained at 4°C. The molecular identification of the isolate was carried out by 16S rRNA gene sequencing. The 16S rRNA gene was amplified using universal primers 518F 5’ccagcagccgcggtaatacg3’ and 800R 5’taccagggtatctaatc3’. PCR products were purified and sequenced. The resultant sequences aligned within the NCBI database (National Centre for Biotechnology Information) using BLASTN.
The phylogenetic tree was constructed using neighbor-joining with Kimura 2-state parameter and pairwise-deletion model analysis implemented in the program MEGA software version X and also evaluated further in a bootstrap analysis of 1,000 replicates. The antimicrobial potential of Nocardiopsis sp. GhM-HA-6 was checked by spot inoculation method using starch agar (10% w/v NaCl, pH 9) (Kumar et al. 2010; Kumar et al. 2018).
The spore suspension of the isolate was spotted on the medium and incubated at 28°C until sporulation. The test organisms, procured from MTCC, Chandigarh (ATCC equivalent), were used to check antimicrobial activity. These included Gram-positive organisms: Bacillus subtilis (MTCC 441), Staphylococcus aureus (MTCC 737), Micrococcus luteus (MTCC 106), Staphylococcus epidermidis (MTCC 3615), Bacillus megaterium (MTCC 428), Bacillus cereus (MTCC 430); Gram-negative organisms: Pseudomonas aeruginosa (MTCC 424), Escherichia coli (MTCC 443), Enterobacter aerogenes (MTCC 8560), Serratia marcescens (MTCC 97), Shigella flexneri (MTCC 1457), Salmonella enterica typhimurium (MTCC 98), Proteus vulgaris (MTCC 1771), Klebsiella pneumonia (MTCC 3384) and three fungi: Aspergillus niger (MTCC 282), Fusarium oxysporum (MTCC 284), Candida albicans (MTCC 227).
The test organisms were grown in nutrient broth at 37○C for 24 hours. The molten nutrient agar, with 1% activated test culture, was poured on the sporulated Nocardiopsis sp. GhM-HA-6. After the incubation of 24 hours at 37○C, the zone of inhibition was measured for each test organism. To achieve the highest production of antimicrobial compounds, effects of various growth conditions were studied by the one variable at a time (OVAT) method.
The effect of various media (starch broth, starch casein broth, actinomycetes broth, glucose asparagine broth, glucose glycine broth, soybean malt broth and complete broth with 10% w/v NaCl and pH 9), effect of starch as a carbon source (0-1.5% w/v), effect of yeast extract as a nitrogenous source (0-1.5% w/v), effect of NaCl (0-15% w/v) and effect of pH (8-11) on the production of antimicrobial compound was checked. All the flasks were incubated at 300C for 7-8 days. Then the cell free filtrates of the culture were collected from all the flasks separately and were tested against the actively growing sensitive culture of S. flexneri by agar well diffusion method.
The isolate was cultivated in starch broth with 10% w/v NaCl and pH 9 on a rotary shaker (120 rpm) at 37○C for 8 days. The cell-free extract was obtained by filtration of broth culture using Whatman No. 1 filter paper. Equal volume of ethyl acetate was added to the culture filtrate for the extraction of the bioactive compounds. The mixture was added to 250 ml glass flask, sealed with a cotton plug, followed by aluminum foil to reduce evaporation of the organic solvent and placed on a shaker for 2 hours at 150 rpm.
Post agitation the mixture was transferred to a separating funnel to generate different layers; the organic layer that contained the secondary metabolites and the aqueous layer. The crude extract was obtained by concentrating the solvent by evaporation and stored at 4○C for further use. Identification of the chemical compounds present in the crude ethyl acetate extract of Nocardiopsis sp. GhM-HA-6 was carried out by Gas Chromatography-Mass Spectrometry (GC-MS) analysis.
Analysis was conducted on a capillary column (Rxi-5ms, 30m, 0.25 mm id, 0.25 µm film thickness) with the following conditions: constant flow of Helium, 1.0 ml min–1; the fixed inlet temperature, 285oC throughout the analysis; injection volume, 3 µl in the linear with an open purge valve (30:1 split ratio); Linear velocity: 36.8 cm/second; Pressure: 65.0 kPa; Purge flow: 3.5 ml/min; Column flow: 1 ml/min; Oven ramp: 80°C holds for 2.0 min, 18°C/min to 260°C holds for 6.0 min, 4°C/min to 285°C holds for 6.0 min; Total run time: 30.25 min. The MS instrument with Ion source temperature: 200°C; Interface temperature: 300°C; Solvent cut time: 5.0 min; Detector voltage: 1 kV; Acquisition mode: Scan mode; Scan speed: 909; Event time: 0.78 second; starting m/z: 40 to 700 m/z. The peaks were identified by comparing the mass spectra with the National Institute of Standards and Technology (NIST, USA) library.
RESULTS AND DISCUSSION
Isolation and Morphology of the Organism: Nocardiopsis sp. GhM-HA-6, a haloalkaliphilic actinomycete, was isolated from the Gulf of Khambhat, Western India. The isolate was characterized on the basis of its cell and colony morphology and Gram’s reaction. The colonies were small sized, round shaped, regular, slightly raised, rough, and opaque. It was Gram-positive, having a filamentous, long thread-like structure. It started sporulation on starch agar after 3 days of incubation with a fluffy mass of spores (Kumar et al. 2018).
Molecular Identification of Nocardiopsis sp. GhM-HA-6 through 16S rRNA Sequencing: The 16S rRNA gene sequencing of the strain Nocardiopsis sp. GhM-HA-6 showed the presence of 1456 bp long 16S rRNA gene in the genomic sequence. The sequence was submitted to NCBI, GenBank, Maryland, USA with accession number (KF384492). The phylogenetic tree was constructed using the neighbor-joining with Kimura 2-state parameter and pairwise-deletion model analysis implemented in the program MEGA software version X and also evaluated in a bootstrap analysis of 1,000 replicates (Figure 1). The molecular characterization through 16S rRNA gene sequencing revealed that the strain belonged to Nocardiopsis sp. (Kumar et al. 2018).
Figure 1: The phylogenetic tree of Nocardiopsis sp. GhM-HA-6
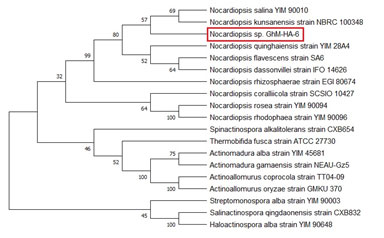
The Antimicrobial Potential: The isolate Nocardiopsis sp. GhM-HA-6 inhibited the growth of five Gram-positive bacteria Bacillus subtilis, Staphylococcus aureus, Micrococcus luteus, Staphylococcus epidermidis, Bacillus cereus, and three Gram-negative bacteria Shigella flexneri, Pseudomonas aeruginosa, Serratia marcescens (Figure 2). The highest inhibition was recorded against Shigella flexneri, however the isolate didn’t show any inhibitory effect against fungi.
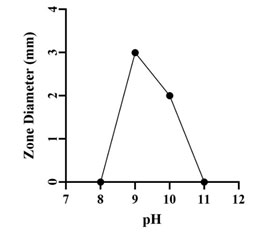
Optimization of Growth Conditions for Antimicrobial Compound Production: Optimum production of the antimicrobial compound, against S. flexneri was obtained in starch broth followed by soybean malt broth, starch casein broth and actinomycetes broth. However, Nocardiopsis sp. GhM-HA-6 wasn’t able to produce any kind of antimicrobial compounds in complete broth, glucose asparagine broth and glucose glycine broth (Figure 3). Optimization of various growth conditions showed that the highest antimicrobial compound production was obtained in the presence of 0.5 % w/v starch, 1% w/v Yeast extract, 10% w/v NaCl and pH 9 (Figure 4-7).
Figure 2: The antibacterial potential of Nocardiopsis sp. GhM-HA-6
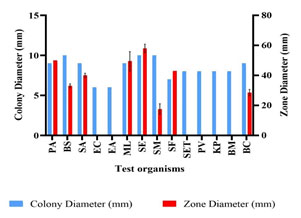
Figure 3: Effect of various media on growth and antimicrobial compounds
production by Nocardiopsis sp. GhM-HA-6 against S. flexneri
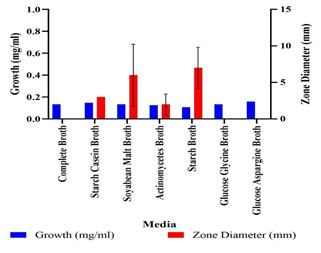
Figure 4: Effect of starch concentrations on antimicrobial compounds
production by Nocardiopsis sp. GhM-HA-6 against S. flexneri
Figure 5: Effect of yeast extract on antimicrobial compounds
production by Nocardiopsis sp. GhM-HA-6 against S.
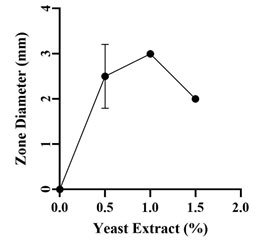
Figure 6: Effect of salt concentrations on antimicrobial compounds
production by Nocardiopsis sp. GhM-HA-6 against S. flexneri
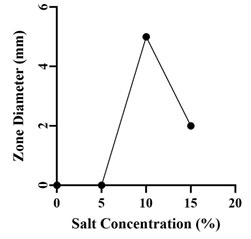
Figure 7: Effect of pH on antimicrobial compounds production by
Nocardiopsis sp. GhM-HA-6 against S. flexneri
Identification of Bioactive Compounds by GC-MS Analysis of Ethyl Acetate Extract: Identification of the bioactive compounds, present in ethyl acetate extract, was carried out using GC-MS analysis. The GC-MS chromatogram of the Nocardiopsis sp. GhM-HA-6 crude extract showed a total of 18 peaks (Figure 8). When compared with the NIST database, the nearest compound hits for those peaks were found (Table 1)
Figure 8: GC-MS chromatogram of Nocardiopsis sp. GhM-HA-6 ethyl acetate extract
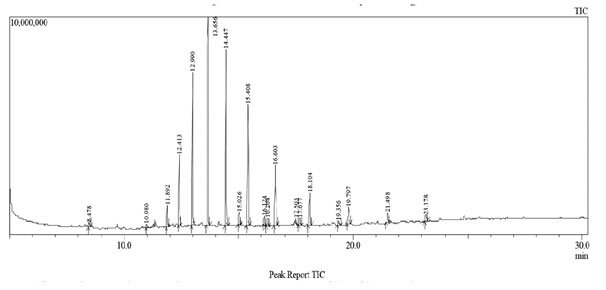
Table 1: GC-MS analysis of Nocardiopsis sp. GhM-HA-6 ethyl acetate extract
| Peak No. | Retention
Time |
Name of the Compound | Molecular Formula | Molecular Weight | Peak Area % |
| 1 | 8.478 | Phenol, 2,4-bis(1,1-dimethylethyl)- | C14H22O | 206.32 | 0.52 |
| 2 | 10.980 | Heptadecane | C17H36 | 240.46 | 1.09 |
| 3 | 11.892 | Heneicosane | C21H44 | 296.6 | 1.76 |
| 4 | 12.413 | Docosane | C22H46 | 310 | 5.85 |
| 5 | 12.990 | Tricosane | C23H48 | 324 | 13.77 |
| 6 | 13.656 | Tetracosane | C24H50 | 338 | 20.94 |
| 7 | 14.447 | Pentacosane | C25H52 | 352 | 18.26 |
| 8 | 15.026 | Hexacosane | C26H54 | 366 | 1.35 |
| 9 | 15.408 | 2-methylpentacosane | C26H54 | 366.7 | 14.08 |
| 10 | 16.124 | Heptacosane | C27H56 | 380 | 0.95 |
| 11 | 16.264 | Tetratetracontane | C44H90 | 619.2 | 1.05 |
| 12 | 16.603 | 2-methylhexacosane | C27H56 | 380.7 | 8.28 |
| 13 | 17.503 | 2,24-dimethylpentacosane | C27H56 | 380.7 | 0.56 |
| 14 | 18.104 | 2-methylheptacosane | C28H58 | 394.8 | 4.78 |
| 15 | 19.356 | Octacosane | C28H58 | 394 | 0.69 |
| 16 | 19.797 | Nonacosane | C29H60 | 408 | 2.52 |
| 17 | 21.498 | Triacontane | C30H62 | 422 | 1.52 |
| 18 | 23.178 | Hentriacontane | C31H64 | 436.85 | 1.01 |
The declining trend in the discovery of new antimicrobial compounds and the enigmatic development of antibiotic resistance in bacteria has increased the need for scientists to search for novel antimicrobial compounds (Genilloud 2017; Durand et al. 2019). Microbial life can be found in the most diverse conditions, including extremes of temperature, pressure, salinity, pH, nutrient concentrations, water availability and presence of high levels of radiations, harmful heavy metals and toxic compounds such as organic solvents and hydrocarbons (Thumar and Singh 2008; Durand et al. 2019Subramani and Sipkema 2019).
Ability to sustain under extremity makes them an interesting system to study the biochemical and molecular basis of adaptations, besides biotechnological aspects (Sharma et al. 2012). In the light of this knowledge, we should explore new habitats in order to identify novel actinobacterial isolates and bioactive compounds. Recently, extremophilic actinobacteria have attracted the researchers with the hope that these organisms would add an inventive dimension to naturally available antimicrobial products research (Zitouni et al. 2004; Vijayakumar et al. 2012; Dhanasekaran et al. 2014; Sharma et al. 2020; Rathore et al. 2021).
Significant studies have not been conducted so far in the Gulf of Khambhat. Therefore, the present study was intended to isolate, screen and characterize antibiotics producing haloalkaliphilic actinomycetes from the Gulf of Khambhat. Nocardiopsis sp. GhM-HA-6, a haloalkaliphilic actinomycete was isolated from the Ghogha site of Bhavnagar district. The isolate was identified as Gram-positive, filamentous, long thread-like structure containing, spore forming actinomycete based on cell and colony morphology and Gram’s staining while molecular characterization confirmed the isolate as Nocardiopsis sp. GhM-HA-6 (Rathore et al. 2021).
The isolate showed the highest similarity with Nocardiopsis salina YIM 90010 (98.46%) and the lowest similarity with Actinomadura gamaensis (90.61%). The isolate was able to inhibit the growth of total eight test organisms. The antimicrobial compounds produced by the isolate were more effective against Gram-positive bacteria as compared to Gram-negative bacteria while the isolate didn’t show any inhibitory effect against fungi. Among all selected test organisms, the highest sensitivity was observed against Shigella flexneri, the intestinal tract pathogen. Sharma et al. (2016) checked the broad-spectrum antimicrobial activity of forest derived soil Actinomycete, Nocardia sp. PB-52 and this study results were quite comparable with it (Sharma et al. 2016; Nithya et al. 2018).
Nithya et al. (2018) isolated 134 morphologically distinct actinobacteria from various soil samples collected from the Saudi Arabian desert. They also used the same methods for isolation, identification, and characterization of antimicrobial compounds from the actinomycete isolates. Range of parameters can affect the production of antimicrobial compounds, so to increase the yield, the effect of growth media, carbon source, nitrogen source, NaCl and pH was checked on the production of antimicrobial compounds by isolate.
Optimization of media showed that the highest antimicrobial compound production and the highest antimicrobial activity against S. flexneri was found in starch broth followed by soybean malt broth, starch casein broth and actinomycetes broth. Optimization showed that the higher production of antimicrobial compounds was observed in media supplemented with complex carbon sources such as starch and malt while production of antimicrobial compounds wasn’t found in media supplemented with glucose like simple carbon source. So, for further production processes starch was used as a carbon source (Elsayed et al. 2020).
The optimization of starch concentration indicated that 0.5% w/v is the optimum concentration required for the highest production of antimicrobial compounds, further increase or decrease in concentration of starch inhibits the production of antimicrobial compounds. Yeast extract was provided as the nitrogen source, initial increase in concentration of yeast extract increased the production of antimicrobial compounds up to 1% w/v, further increase in yeast extract concentration decreased the production.
The isolate showed the highest production of antimicrobial compounds at 10% w/v NaCl followed by 15 % w/v NaCl. The isolate wasn’t able to produce antimicrobial compounds in absence or at lower concentrations of NaCl. Optimization of pH showed that the isolate was able to produce antimicrobial compounds at pH 9 and 10 but the highest activity was observed at pH 9 (Kumar et al. 2019).
The GC-MS chromatogram of the Nocardiopsis sp. GhM-HA-6 ethyl acetate extract indicated the presence of 18 compounds, from which 15 compounds have already been reported to have antimicrobial potential against variety of pathogens including Phenol, 2, 4-bis (1, 1-dimethylethyl); Heptadecane; Heneicosane; Docosane, Tricosane, Tetracosane, Pentacosane, Hexacosane; Heptacosane; Tetratetracontane; 2-methylhexacosane; Octacosane; Nonacosane; Triacontane; Hentriacontane while methyl derivatives of alkanes identified from ethyl acetate extract haven’t been reported for antimicrobial activity (Padmavathi et al. 2014; Nandhini et al. 2015; Begum et al. 2016; El-Naggar et al. 2017; Kumar et al. 2019; Elsayed et al. 2020).
CONCLUSION
The findings of the present study indicated that Nocardiopsis sp. GhM-HA-6, isolated from the Ghogha site, inhibited the growth of various test organisms including Staphylococcus epidermidis, Pseudomonas aeruginosa, Shigella flexneri, and Staphylococcus aureus. The ethyl acetate extract of Nocardiopsis sp. GhM-HA-6, indicated the presence of potent antimicrobial compounds such as Phenol-2, 4-bis (1, 1-dimethylethyl) – which can further be purified and used for the treatment of various known and unknown infections. In nutshell, our findings in this field will surely be a significant contribution to the knowledge of compounds unique from marine bacteria as potential sources of new drugs in the pharmacological industry.
ACKNOWLEDGEMENT
This study was financially supported by the Department of Biotechnology (New Delhi, India) (DBT Sanction Order No.: BT/Bio-CARe/03/596/2010-11).
Conflict of Interest: Authors declare no conflicts of interests to disclose.
REFERENCES
Becerril-Espinosa A, Freel KC, Jensen, PR et al. (2013) Marine Actinobacteria from the Gulf of California: diversity, abundance and secondary metabolite biosynthetic potential Antonie van Leeuwenhoek Vol 103 Page 809–819. https://doi.org/10.1007/s10482-012-9863-3
Bennur T, Kumar AR, Zinjarde S et al. (2015) Nocardiopsis species: Incidence, ecological roles and adaptations Microbiological Research Vol 174 Page 33–47 https://doi.org/10.1016/j.micres.2015.03.010
Chakrabarti T (1998) ACTINOMYCETES – Isolation, Screening, Identification, and Gene Cloning in Streptomyces Laboratory manual. MTCC, Institute of Microbial Technology, Chandigarh.
Cook AE and Meyers PR (2003) Rapid identification of filamentous actinomycetes to the genus level using genus-specific 16S rRNA gene restriction fragment patterns Int J Syst Evol Microbiol, Vol 53 No 6 Page 1907-15. doi: https://doi.org/10.1099/ijs.0.02680-0 PMID: 14657122.
Dhakal D, Pokhrel AR, Shrestha B et al. (2017) Marine rare actinobacteria: isolation, characterization, and strategies for harnessing bioactive compounds Front Microbiol Vol 8 Page 1106. https:// doi.org/10.3389/fmicb.2017.01106
Dhanasekaran D, Thajuddin N, Panneerselvam A et al. (2014) Isolation, Characterization of Antibacterial Methyl Substituted β-Lactam Compound from Streptomyces noursei DPTD21 in Saltpan Soil, India TBAP Vol 4 No 2 Page 71 – 88.
Dolak LA, Castle TM and Laborde AL (1980) 3-Trehalosamine, a new disaccharide antibiotic J Antibiot Vol 7 Page 690–4.
Durand GA, Raoult D and Dubourg G (2019) Antibiotic discovery: history, methods and perspectives International Journal of Antimicrobial Agents Vol 53 No 4 Page 371–382.
El-Naggar NE, El-Bindary AA, Abdel-Mogib M et al. (2017) In vitro activity, extraction, separation and structure elucidation of antibiotic produced by Streptomyces anulatus NEAE-94 active against multi-drug resistant Staphylococcus aureus Biotechnology & Biotechnological Equipment Vol 31 No 2 Page 418-430, DOI: https://doi.org/10.1080/13102818.2016.1276412
Elsayed TR, Galil DF, Sedik MZ, et al. (2020) Antimicrobial and Anticancer Activities of Actinomycetes Isolated from Egyptian Soils, Int. J. Curr. Microbiol. App. Sci Vol 9 No 9 Page 1689-1700.
Engelhardt K, Degnes KF, Kemmler M, et al. (2010a) Production of a new thiopeptide antibiotic TP-1161, by a marine Nocardiopsis species Appl Environ Microbiol Vol 76 Page 4969–76.
Begum FI, Mohankumar R, Jeevan M and Ramani K (2016) GC-MS Analysis of Bio-active Molecules Derived from Paracoccus pantotrophus FMR19 and the Antimicrobial Activity Against Bacterial Pathogens and MDROs Indian J Microbiol Vol 56 No 4 Page 426-432. doi: https://doi.org/10.1007/s12088-016-0609-1
Genilloud O (2017) Actinomycetes: still a source of novel antibiotics Natural Product Reports, Vol 34 No 10 Page 1203–1232.
Ibrahim AH, Desoukey SY, Fouad MA, et al. (2018) Natural Product Potential of the Genus Nocardiopsis Marine Drugs Vol 16 No 5 Page 147. https://doi.org/10.3390/md16050147
Jose PA and Jebakumar SR (2012) Phylogenetic diversity of actinomycetes cultured from coastal multipond solar saltern in Tuticorin, India Aquat Biosyst. Vol 8 Page 23. doi:10.1186/2046-9063-8-23
Kamjam M, Sivalingam P, Deng Z et al. (2017) Deep Sea Actinomycetes and Their Secondary Metabolites Front. Microbiol Vol 8 Page 760. doi: 10.3389/fmicb.2017.00760
Kroppenstedt RM and Evtushenko LI (2006) The family Nocardiopsaceae. The Prokaryotes: A Handbook on the Biology of Bacteria Springer-Verlag 3(3) 754–95.
Kumar N, Menghani E and Mithal R (2019) GCMS analysis of compounds extracted from Actinomycetes AIA6 isolates and study of its antimicrobial efficacy Indian Journal of Chemical Technology 26 362-370.
Kumar N, Singh RK, Mishra SK, et al. (2010) Isolation and screening of soil Actinomycetes as source of antibiotics active against bacteria International Journal of Microbiology Research 2(2) Page 12-16.
Kumar PS, Duraipandiyan V and Ignacimuthu S (2014) Isolation, screening and partial purification of antimicrobial antibiotics from soil Streptomyces sp. SCA 7 Kaohsiung Journal of Medical Sciences 30(9) Page 435-446.
Kumar S, Stecher G, Li M, et al. (2018) MEGA X: Molecular Evolutionary Genetics Analysis across Computing Platforms Mol. Biol. Evol. 35 Page 1547–1549.
Meklat A, Sabaou N, Zitouni A, et al. (2011) Isolation, taxonomy, and antagonistic properties of halophilic actinomycetes in Saharan soils of Algeria Appl Environ Microbiol 77 Page 6710–4.
Meyer J (1976) Nocardiopsis, a new genus of the order Actinomycetales Int J Syst Bacteriol 26 Page 487–93.
Mwirichia R, Muigai AW, Tindall B, et al. (2010) Isolation and characterization of bacteria from the haloalkaline Lake Elmenteita, Kenya Extremophiles 14 Page 339–48.
Nandhini US, Sangareshwari S and Lata K (2015) Gas chromatography-mass spectrometry analysis of bioactive constituents from the marine Streptomyces Asian J Pharm Clin Res 8 Page 244–246.
Nithya K, Muthukumar C, Biswas B, et al. (2018) Desert actinobacteria as a source of bioactive compounds production with a special emphasis on Pyridine-2, 5-diacetamide a new pyridine alkaloid produced by Streptomyces sp. DA3-7 Microbiological Research 207 Page 116–133.
Padmavathi AR, Bose A and Pandian SK (2014) Phenol, 2, 4-bis (1, 1-dimethylethyl) of marine bacterial origin inhibits quorum sensing mediated biofilm formation in the uropathogen Serratia marcescens Biofouling 30(9) Page 1111–1122, https://doi.org/10.1080/08927014.2014.972386
Rainey FA, Rainey NW, Kroppenstedt RM et al. (1996) The Genus Nocardiopsis represents a phylogenetically coherent taxon and a distinct Actinomycete lineage: proposal of Nocardiomaceae fam. nov. Int J Syst Bacteriol 46 Page 1088–92.
Ramnathan V, Vincent D, Sundrmoorthy S, et al. (2002) Critical habitat information system for Gulf of Khambhat-Gujarat; Govt. of India: Department of Ocean development.
Rathore DR, Sheikh M, Gohel GD et al. (2021) Genetic and phenotypic heterogeneity of the Nocardiopsis alba strains of sea water Current Microbiology 78 Page 1377-1387.
Romanenko LA, Tanaka N, Kalinovskaya NI et al. (2013) Antimicrobial potential of deep surface sediment associated bacteria from the Sea of Japan World J Microbiol Biotechnol 29 Page 1169–77.
Roy S, Chandni S, Das I, et al. (2015) Aquatic model for engine oil degradation by rhamnolipid producing Nocardiopsis VITSISB Biotech 5 Page 153–164. doi: https://doi.org/10.1007/s13205-014-0199-8
Schorn MA, Alanjary MM, Aguinaldo K, et al. (2016) Sequencing rare marine actinomycete genomes reveals high density of unique natural product biosynthetic gene clusters Microbiology 162 Page 2075–2086.
Sharma AK, Gohel S and Singh SP (2015) Actinobase: Database on molecular diversity, phylogeny and biocatalytic potential of salt tolerant alkaliphilic actinomycetes Bioinformation Vol 8 No 11 Page 535-538. DOI: https://doi.org/10.6026/97320630008535
Sharma AK, Kikani BA and Singh SP (2020) Biochemical, thermodynamic and structural characteristics of a biotechnologically compatible alkaline protease from a haloalkaliphilic, Nocardiopsis dassonvillei OK-18 International Journal of Biological Macromolecules (IJBIOMAC) 153 Page 680-696.
Sharma P, Kalita MC and Thakur D (2015) Broad Spectrum Antimicrobial Activity of Forest-Derived Soil Actinomycete, Nocardia sp. PB-52 Frontiers in Microbiology 7 Page 347. https://doi.org/10.3389/fmicb.2016.00347
Sivakumar K, Sahu MK, Thangaradjou T et al. (2007) Research on marine actinobacteria in India Indian J. Microbiol. 47 Page 186-196 https://doi.org/10.1007/s12088-007-0039-1
Solanki D, Kanejiya J, Beleem I et al. (2016) Checklist of intertidal marine fauna in mangrove ecosystem, Ghogha coast, Gulf of Khambhat, India Journal of Entomology and Zoology Studies 4(4) Page 1281-1284.
Subramani R and Sipkema D (2019) Marine Rare Actinomycetes: A Promising Source of Structurally Diverse and Unique Novel Natural Products Mar. Drugs 17 Page 249 doi:10.3390/md17050249
Takahashi A, Hotta K, Saito N, et al. (1986) Production of novel antibiotic, dopsisamine, by a new subspecies of Nocardiopsis mutabilis with multiple antibiotic resistance J Antibiotics 39 Page 175–83.
Thumar JT and Singh SP (2008) Organic solvent tolerance of an alkaline protease from salt-tolerant alkaliphilic Streptomyces clavuligerus strain Mit-1 J Ind Microbiol Biotechnol 6 Page 211–218. DOI https://doi.org/10.1007/s10295-008-0487-6
Vijayakumar R, Panneerselvam K, Muthukumar C, et al. (2012) Antimicrobial potentiality of a halophilic strain of Streptomyces sp. VPTSA18 isolated from the saltpan environment of Vedaranyam, India Ann. Microbiol. 62 Page 1039–1047.
Wu ZC, Li S, Nam SJ, et al. (2013) Nocardiamides A and B, two cyclohexapeptides from the marine-derived actinomycete Nocardiopsis sp. CNX037 J Nat Prod 76 Page 694–701.
Zitouni A, Boudjella H, Lamari L, et al. (2005) Nocardiopsis and Saccharothrix genera in Saharan soils in Algeria: isolation, biological activities and partial characterization of antibiotics Res Microbiol 156 Page 984–93.
Zitouni A, Lamari L, Boudjella H, et al. (2004) Saccharothrix algeriensis sp. nov.: isolated from Saharan soil Int. J. Sys. Evol. Microbiol 54 Page 1377–1381.


