1Department of Biological Sciences, Faculty of Science, King Abdulaziz University, 21589 Jeddah, SAUDI ARABIA
2Department of Medical Laboratory Sciences, Faculty of Applied Medical Sciences, Shaqra, Saudi Arabia
3Princess Doctor Najla Bint Saud Al Saud Distinguished Research Centre For Biotechnology, Jeddah, Saudi Arabia
4Department Of Genetics, Faculty Of Agriculture Ain Shams University, Cairo, Egypt
5Center Of Excellence In Bionanoscience Research, King Abdulaziz University, 21589 Jeddah, Saudi Arabia
6King Abdul Aziz University, Faculty of Science, Biochemistry Department, Jeddah, P.O. Box 40288, zip code 21499.
Article Publishing History
Received: 14/10/2020
Accepted After Revision: 20/12/2020
The widespread and a dramatic increase usage of antibiotics in recent years led to appearance of new bacterial resistance to a various classes of antibiotics. The bacterial resistance to different antibiotics leads to increased morbidity and mortality associated with multidrug-resistant (MDR), particularly in pathogenic bacteria that infect people and cause infections. The current study was aimed to study and evaluate the resistance of some chosen Gram positive bacterial strains, it were Bacillus cereus, Enterococcus faecalis, Staphylococcus aureus and Staphylococcus saprophyticus isolated from King Fahad Hospital, Jeddah, Saudi Arabia to 16 different chosen antibiotics. The evaluation was done through disk diffusion assay method. Results concluded that the bacterial strains under study were resistant to all used antibiotics. This main point of study was confirmed by the presence of antibiotic resistance genes i.e. ampR, blashv-12, penA1, penA2, ampC1, ampC2, aac4A-cr, and vanR on plasmid and genomics DNA isolated from used bacterial strains. The obtained results highlights that the increase and rapid used of antibiotic resistance lead to appearance of MDR strains
Antibiotic Resistance, Pathogenic Bacteria, DNA, Plasmids, Genes.
Omar A. S, Alharthi M. S. F, Al-Saud N. S, Abo-Aba S. E. M, Omar U. M, Hassan S. M. Molecular Studies of Antibiotic Resistant genes in some Gram-Positive Bacteria Isolated from King Fahad Hospital, Jeddah, Saudi Arabia. Biosc.Biotech.Res.Comm. 2020;13(4).
Omar A. S, Alharthi M. S. F, Al-Saud N. S, Abo-Aba S. E. M, Omar U. M, Hassan S. M. Molecular Studies of Antibiotic Resistant genes in some Gram-Positive Bacteria Isolated from King Fahad Hospital, Jeddah, Saudi Arabia. Biosc.Biotech.Res.Comm. 2020;13(4). Available from: https://bit.ly/3nAIG3X
Copyright © Omar et al., This is an Open Access Article distributed under the Terms of the Creative Commons Attribution License (CC-BY) https://creativecommons.org/licenses/by/4.0/, which permits unrestricted use distribution and reproduction in any medium, provide the original author and source are credited.
INTRODUCTION
Bacteria are leading causes for infectious diseases in humans. . Gram-positive bacteria’s are considered as main causative agents for animal intestinal and potentially lethal foodborne diseases in humans (Songer 2010; Maurya and Agarwal, 2019).One among the class of gram positive bacteria’s are Bacillus cereus and is common in nature. . This bacterium is isolated from plants soil, and intestinal tracks of some mammals and insects. From these habitats, there are many sources by which it is spreader and are responsible for an emetic or a diarrhoeal type of food-related (Arnesen et al., 2008). Another important class of these bacteria’s are Enterococci which are found in human faecal flora and are main causative agent for the Nasocomial infections in humans. thr . Human Enterococcal infections are caused by two species, Enterococcus faecalis and Enterococcus faecium (Lebreton et al., 2014).
Antimicrobial resistance is a growing public health threat worldwide and it is due to massive imprudent usage of antibiotics in clinical practice that is responsible for resistance in bacterial species. (Livermore, 2003; Tenover, 2006; Kapoor et al., 2017). There is a global concern about the appearance and growth of bacterial resistance to commonly used antibiotics (Cizman, 2003; Maurya and Agarwal, 2019). Some pathogenic bacteria developed resistance to multiple classes of antibiotics. The loss of effective antibiotics will undermine the management of the common infectious complications in susceptible patients undergoing chemotherapy for cancer, dialysis for renal failure and surgery, especially organ transplantation, for which the ability to treat secondary infections is crucial and even our ability to fight bacterial diseases (Tanwar et al., 2014).
There are various reasons that led to the spread of antibiotic resistance and possibly the most important cause is the non-application of infection control within the hospitals and outside. It has been recognized that methicillin-resistant Staphylococcus aureus (MRSA) in a hospital and in the community are genetically related. Another reason is the use of antibiotics in non-human outlets. A combination of microbial properties, the selective pressure of antibiotic use, communal and technical changes that enhance the transmission of resistant organisms has been attributed to the increase in bacterial resistance to antibiotics (Borer et al., 2002). Many conjugative transposons and plasmids, integrons and bacteriophages discovered to date encode antibiotic resistance proteins and can be transferred from donor bacteria to recipient bacteria via horizontal gene transfer.
Resistance could be acquired also by a mutation in the genes in the. Resistance mechanisms probably have developed from genes present in organisms that produce antibiotics (Poole, 2002; Tanwar et al., 2014). Further some small bacterial regulatory RNAs, categorized as RNA attenuators and small RNAs, coud control the expression of some resistance genes. For example, small RNA sprX, can formulate bacterial resistance to glycopeptide antibiotics through the specific downregulation of SpoVG protein. Modifications of the target enzymes or antibiotics can prevent the binding to targets, which act to confer high levels of resistance in respiratory/oral bacteria (Jiang et al., 2018).
Resistant bacteria can be transmitted to humans through direct contact and via food chain or indirectly from the ecological pollution of farm effluents. Therefore, the current study was aimed to re-evaluate the antibiotic-resistance statues for some Gram-positive by the molecular characterization of these bacteria. Our PCR results from both genomics and plasmid DNA confirmed presence of resistant genes for different antibiotics in these bacterial strains.
MATERIAL AND METHODS
Bacterial Strains: Following Gram-positive were used for this study (Bacillus cereus, Enterococcus faecalis, Staphylococcus aureus and Staphylococcus saprophyticus). And all the strains were kindly obtained from King Fahad Hospital, Jeddah, Saudi Arabia.
Antibiotic susceptibility testing rings: MASTRING-ST obtained from Mast Group Ltd. Bootle, Merseyside, U.K. an antibiotic susceptibility ring device for the convenient, simultaneous testing of the equivalent of six antibiotic discs as shown in supplementary Table (1).
Preparation of bacterial suspension: Using a sterile loop, three discrete colonies were taken from a 24 hour culture of the the bacteria and were inoculated into 3 ml of nutrient broth. The broth was incubated for 2-3 hours until a slight visible turbidity appears. The growth of the bacteria was checked by the spectrophotometer at absorbance at 630 nm.
Performing the disk-diffusion susceptibility test: A sterile cotton swab was dipped into the standardized suspension, and excess broth was expressed by pressing and rotating the swab firmly against the inside of the tube above the fluid level. To inoculate the agar plates, the swab was then streaked evenly in three directions over the entire surface of the Mueller-Hinton agar plate to obtain a uniform inoculum. A final sweep was made of the agar rim with the cotton swab. This plate was then allowed to dry for 3 to 5 minutes.
The antibiotic impregnated discs were applied to the surface of the inoculated plate, using sterile forceps. All disks were gently pressed down into the agar with the forceps to ensure complete contact with the agar surface and incubated at 37 C for 20 hours as per Kiser et al., 2011. Primer design The sequences of antibiotic resistance genes were obtained from GeneBank nucleotide databases. The primers for each antibiotics resistant gene were designed by primer3 program The names of antibiotic resistant genes, their accession numbers, primer sequences and Sizes of RT-PCR Products as shown in Table (1).
Table 1. Resistance and susceptibility patterns of some Gram-positive bacteria to some antibiotics using disk-diffusion susceptibility test
| Antibiotic Name | Bacteria/Diameter of inhibition zone in mm | |||
| Bacillus cereus | Enterococcus faecalis | Staphylococcus aureus | Staphylococcus saprophyticus | |
| Amikacin (AK) | 20 | R | 40 | 23 |
| Gentamicin (GM) | 20 | 20 | 34 | 25 |
| Ampicillin (AP) | R | R | R | R |
| Augmentin (AUG) | R | R | R | 18 |
| Aztreonam (ATM) | R | R | R | R |
| Cefoxitin (FOX) | R | R | R | R |
| Ceftazidime (CAZ) | R | R | R | R |
| Cephalothin (KF) | R | R | 50 | R |
| Ciprofloxacin (CIP) | 20 | 74 | 22 | 16 |
| Imipenem (IMI) | 36 | 77 | 40 | 46 |
| Meropenem (MEM) | R | R | R | R |
| Penicillin (P) | R | R | R | R |
| Piperacillin (PRL) | 16 | 20 | R | 14 |
| Trimethoprim/Sulfamethoxazole (TS | R | R | 40 | 75 |
| Vancomycin (VA) | 22 | R | 10 | 32 |
| Ampicillin (AP) | R | R | R | R |
| Nalidixic acid (NA) | 20 | 30 | 30 | 22 |
Plasmid and genomic DNA Extraction Protocol: The plasmid and genomic DNA extraction was carried out by plasmid extraction mini prep kit and Thermo Scientific GeneJET Chromosomal Purification Kit respectively (Thermo Scientific GeneJET) according to according to manufacturer’s instruction.
Polymerase chain reaction (PCR):All samples were placed in a polymerase chain reaction program, the PCR program was run as the follows, 40 cycles at 95°C for 4 min, 95°C for 15 s, 50-60°C for 18 s, 72°C for 30 s and a final extension at 72°C for 7 minutes. The PCR products were analyzed on a 2% agarose gel stained with 5 µl Ethidium Bromide (EtBr). Bands on the gel were visualized in gel documentation system.
RESULTS AND DISCUSSION
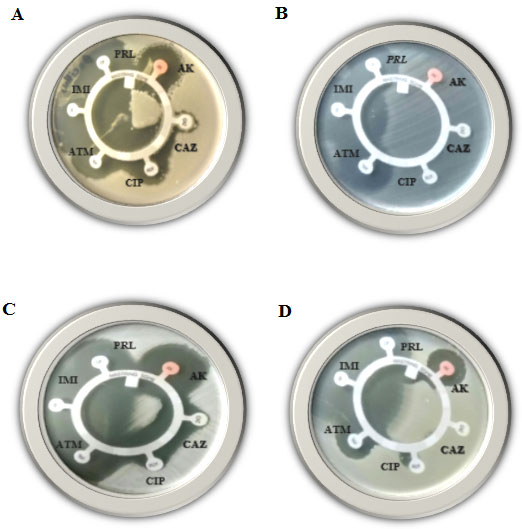
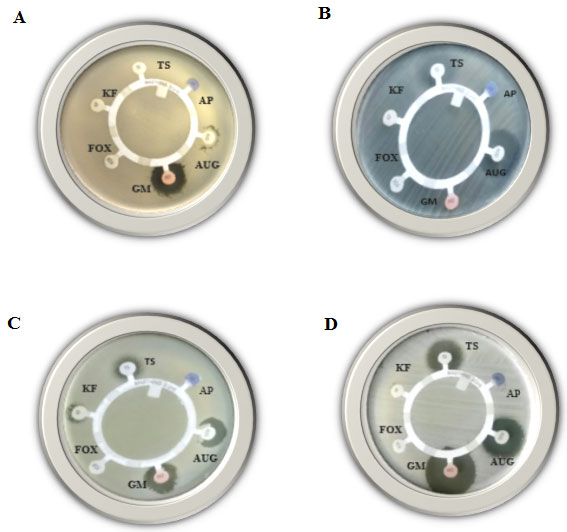
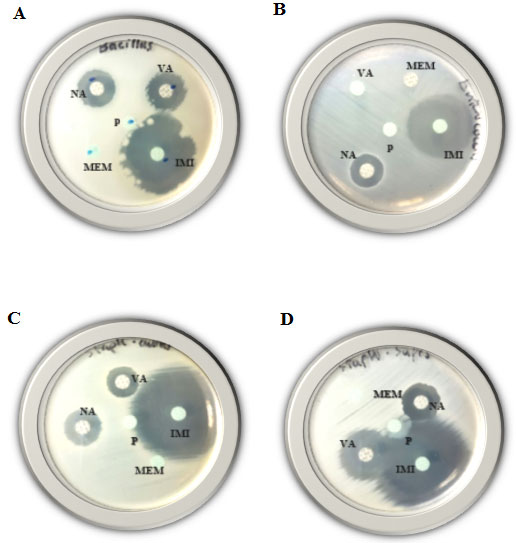
Disk-diffusion susceptibility results: In the present study, the four mentioned Gram-positive species were examined for their resistance against 16 different antibiotics using disk-diffusion susceptibility test. The inhibition zone diameter chart was as follows; resistant: 13 mm or less, intermediate: 14-16 mm, sensitive: 17 mm or more. If present, the diameter of the inhibition zones in the current study ranged from 14 -77 mm.As shown (Table, 3) and Figures (1, 2 & 3) all mentioned species revealed resistance to Ampicillin, Aztreonam, Ceftazidime, Meropenem and Penicillin, while all of them found to be resistant to Augmentin and Cefoxitin antibiotics except Staphylococcus saprophytic, which showed sensitivity to both. As well as all of them were resistant to Cephalothin antibiotic except Staphylococcus aureus. Bacillus cereus and Enterococcus faecalis both were resistant to Sulfadiazine/trimethoprim antibiotic. On the other hand, three species showed resistance to only on antibiotic, Staphylococcus aureus, Enterococcus faecalis and Enterococcus faecalis found to be resistant to Piperacillin, Amikacin and Vancomycin antibiotics, respectively.
Figure 1: Disk-diffusion susceptibility test for the Gram-positive bacteria; Bacillus cereus (A), Enterococcus faecalis (B), Staphylococcus aureus (C) and Staphylococcus saprophyticus (D), using AK, CAZ, CIP, ATM, IMI and PRL antibiotics.
Figure 2: Disk-diffusion susceptibility test for the Gram-positive bacteria; Bacillus cereus (A), Enterococcus faecalis (B), Staphylococcus aureus (C) and Staphylococcus saprophyticus (D), using AP, AUG, GM, FOX, KF and TS antibiotics.
Figure 3: Disk-diffusion susceptibility test for the Gram-positive bacteria; Bacillus cereus (A), Enterococcus faecalis (B), Staphylococcus aureus (C) and Staphylococcus saprophyticus (D) using IMI, MEM, VA, NA and P antibiotics.
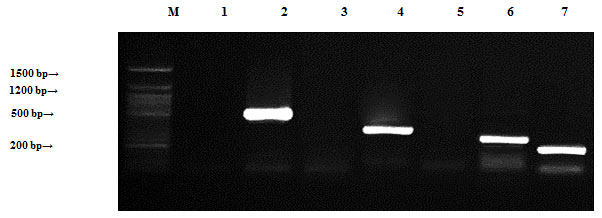
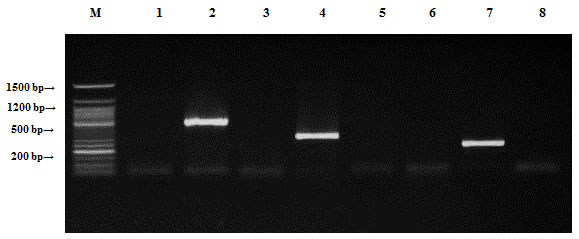
Localization of antibiotic resistance genes:All plasmids and genomic DNA isolated from the four mentioned species were separately tested by PCR to determine the presence of antibiotic resistance genes.Bacillus cereus samples were tested by PCR to determine the presence of antibiotic resistance genes (ampR, blashv-12, penA1, ampC1, aac4A-cr, ampC2 and penA2). Plasmids found to harbor four out of the six tested antibiotic resistance genes (blashv-12, ampC1, aac4A-cr and penA2) with molecular sizes of 491, 323, 296 and 235 bp, respectively (Figure, 4). Chromosomal DNA harbored four out of the six tested antibiotic resistance genes (ampR, blashv-12, ampC1 and penA2) with molecular sizes of 310, 491, 323 and 235 bp, respectively (Figure, 5).
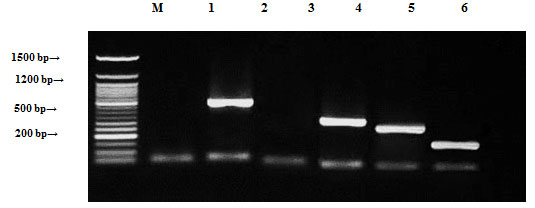
Enterococcus faecalis samples were tested by PCR to confirm the presence of ampR, blashv-12, penA1, ampC1, aac4A-cr, ampC2, penA2 and vanR. Plasmids found to harbor four out of the eight tested genes blashv-12, ampC1, aac4A-cr and ampC2, with molecular sizes of 491, 323, 226 and 296 bp, respectively, as shown in Figure (6). Chromosomal DNA found to harbor four out of the eight tested antibiotic resistance genes (blashv-12, ampC1 and penA2) with molecular sizes of 491, 323 and 235 bp, respectively, as shown in Figure (7).
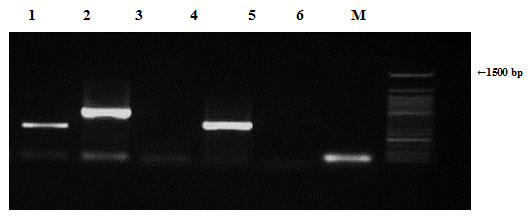
Staphylococcus aureus samples were also tested by PCR to determine the presence of antibiotic resistance genes (ampR, blashv-12, penA1, ampC1, ampC2 and penA2). Plasmids harbored four out of the six tested antibiotic resistance genes (blashv-12, ampC1, ampC2 and penA2) with molecular sizes of 491, 323, 296 and 235 bp, respectively, as shown in Figure (8). Chromosomal DNA found to harbor four out of the six tested antibiotic resistance genes (ampR, blashv-12, and ampC1) with molecular sizes of 310, 491, and 323 bp, respectively, as shown in Figure (9).
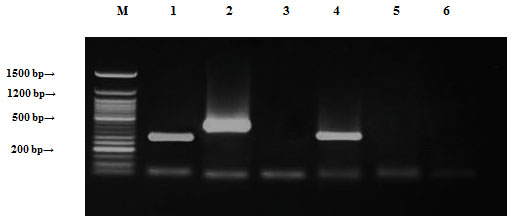
Staphylococcus saprophyticus samples were tested by PCR to decide the presence of genes ampR, blashv-12, penA, ampC1, ampC2 and penA2 genes. It found to harbor three out of the six tested antibiotic resistance genes (ampR, blashv-12 and aac4A-cr) with molecular sizes of 310, 491 and 226 bp, respectively, as shown in Figure (10). Chromosomal DNA found to harbor four out of the six tested antibiotic resistance genes (blashv-12, ampC1 and penA2) with molecular sizes of 491, 323 and 235 bp, respectively, as shown in Figure (11).
Figure 4: Detection of Bacillus cereus antibiotic resistance genes of DNA plasmids using agarose gel electrophoresis of the PCR products. Lane 1, ampR; 2, blashv-12; 3, penA1; 4, ampC1; 5, aac4A-cr; 6, ampC2; 7, penA2 and M refers to 50 bp DNA ladder.
Figure 5: Detection of Bacillus cereus antibiotic resistance genes of chromosomal DNA using agarose gel electrophoresis of the PCR products. Lane 1, ampR; 2, blashv-12; 3, penA1; 4, ampC1; 5, aac4A-cr; 6, ampC2; 7, penA2 and M refers to 50 bp DNA ladder.
Figure 6: Detection of Enterococcus faecalis antibiotic resistance genes of plasmid DNA using agarose gel electrophoresis of the PCR products. Lane 1, ampR; 2, blashv-12; 3, penA1; 4, ampC1; 5, aac4A-cr; 6, ampC2; 7, penA2; 8, vanR and M refers to 50 bp DNA ladder.
Figure 7: Detection of Enterococcus faecalis antibiotic resistance genes of chromosomal using agarose gel electrophoresis of the PCR products. Lane 1, ampR; 2, blashv-12; 3, penA1; 4, ampC1; 5, aac4A-cr; 6, ampC2; 7, penA2; 8, vanR and M refers to 50 bp DNA.
Figure 8: Detection of Staphylococcus aureus antibiotic resistance genes of DNA plasmids using agarose gel electrophoresis of the PCR products. Lane 1, ampR; 2, blashv-12, 3, penA1; 4, ampC1; 5, ampC2; 6, penA2 and M refers to 50 bp DNA ladder.
Figure 9: Detection of Staphylococcus aureus antibiotic resistance genes of chromosomal DNA using agarose gel electrophoresis of the PCR products. Lane 1, ampR; 2, blashv-12, 3, penA1; 4, ampC1; 5, ampC2; 6, penA2 and M refers to 50 bp DNA ladder.
Figure 10: Detection of Staphylococcus saprophyticus antibiotic resistance genes of DNA plasmids using agarose gel electrophoresis of the PCR products. Lane 1, ampR; 2, blashv-12; 3, penA1; 4, ampC1; 5, ampC2; 6, penA2 and M refers to 50 bp DNA ladder.
Figure 11: Detection of Staphylococcus saprophyticus antibiotic resistance genes of chromosomal DNA using agarose gel electrophoresis of the PCR products. Lane 1, ampR; 2, blashv-12; 3, penA1; 4, ampC1; 5, ampC2; 6, penA2 and M refers to 50 bp DNA ladder.
Gram-positive bacteria are the most common bacterial pathogens that cause diseases in humans, with streptococci and staphylococci occurring most frequently. Antibiotic resistance among Gram-positive bacteria has been increasing steadily during the past several decades. (Turutoglu et al., 2006). In the current study, Bacillus cereus found to be sensitive to two out of the ten used β-lactam antibiotics (Imipenem and Piperacillin), while was resistant to eight of them. It also found to be sensitive to Aminoglycoside (Amikacin and Gentamicin), sensitive to Quinolones (Ciprofloxacin and Nalidixic acid), resistant to Trimethoprim/Sulfamethoxazole as member of Sulfonamides group and sensitive to Vancomycin from Glycopeptides group.
A previous study by Whitman et al. (1998) reported that, all Bacillus cereus strains was susceptible to Imipenem, Vancomycin, Gentamicin and Ciprofloxacin. They also reported that, Bacillus cereus was rarely susceptible to Penicillin. Bacillus cereus is usually sensitive to Aminoglycosides and Vancomycin. Part of our findings are in accordance with the findings of a previous study (Leach et al., 2007). On the other hand, in contrast to the same mentioned previous study, our results revealed that, Bacillus cereus has acquired resistance to new antibiotics, which it was sensitive to before. This increased resistance might came from acquiring resistance genes from other bacterial species via horizontal gene transfer by time.
Our results showed that, Enterococcus faecalis was resistant to most of the tested β-lactam antibiotics except for Imipenem and Piperacillin. It also found to be resistant to Amikacin as an Aminoglycoside, Trimethoprim/Sulfamethoxazole (Sulfonamides) and Vancomycin (Glycopeptides). This is consistent with another study, which reported that, Enterococcus faecalis was resistant to Ampicillin, Penicillin, and Gentamicin, while the difference found is the resistance of Enterococcus faecalis to Ciprofloxacin (Mengeloğlu et al., 2011).The present study results revealed Staphylococcus aureus as a resistant species to some β-lactams such as Ampicillin, Augmentin, Aztreonam, Cefoxitin, Ceftazidime, Meropenem, Penicillin and Piperacillin. This results are consistent with previous studies that proved Staphylococcus aureus is resistant to Penicillin, β-lactams and Cephalosporins (McDougal and Thornsberry, 1986; Turutoglu et al., 2006).
Staphylococcus saprophyticus resisted most of the tested β-lactam antibiotics (Ampicillin, Aztreonam, Ceftazidime, Cephalothin, Meropenem and Penicillin), these results are in accordance with the results of another study, which reported that, Staphylococcus saprophytic was resistant to most of β-lactam antibiotics (Ferreira et al., 2013). In the present study, none of our Staphylococci species was resistant to vancomycin. Vancomycin has long been considered as an antibiotic of last resort for MDR staphylococci infections (Koksal et al., 2009). On the other hand, vancomycin resistance has developed first in enterococci, in Staphylococcus aureus and coagulase-negative staphylococci (Boneca and Chiosis, 2003, Palazzo et al., 2005).
This situation has led Staphylococci to become a serious health problem that medical practitioners should be concerned about. The extensive use of glycopeptides in hospitals has been related to decreased susceptibility to these agents. Unfortunately, the therapy chance of MDR staphylococci infections is gradually decreasing. Molecular genetic analysis by PCR technique using plasmids and chromosomal DNA isolated from the four tested species revealed that, they harbor some but not all of the β-lactam resistant genes, which they are resistant to their corresponding antibiotics. This result is acceptable because as previously mentioned (Thenmozhi et al., 2014), the extended spectrum β-lactamases (ESBL) are enzymes produced by a variety of Gram-negative bacteria, which confer an increased resistance to commonly used antibiotics.
In conclusion, some species harbor some of the resistance genes on the plasmids only, some have them on their chromosomal DNA only and others harbor them on both plasmids and chromosomes. Together, these findings further supported by the facts that, horizontal gene transfer or the process of swapping genetic material between neighboring “contemporary” bacteria, is another means by which resistance can be acquired. Many of the antibiotic resistance genes are carried on plasmids, transposons or integrons that can act as vectors that transfer these genes to other members of the same bacterial species, as well as to bacteria in another genus or species. Horizontal gene transfer may occur via three main mechanisms: transformation, transduction or conjugation. Acquired resistance genes can be further incorporated into the recipient chromosome by recombination (Roberts, 2005, Von Wintersdorff et al., 2016).
Although antibiotic resistance occurs naturally, overuse and misuse of antibiotics in humans and animals is accelerating the process. Steps can be taken at all levels of society to reduce the impact and limit the spread of resistance, including the public, who can help by preventing infection through good hygiene and vaccination, only using antibiotics when prescribed by a certified health professional, taking the full course and never sharing or using leftover antibiotics. Therefore, the current study shows the importance of monitoring antibiotic intake and resistance trends of nosocomial infections, especially with infection control measures, to prevent the occurrence and blowout of multi-drug resistant bacteria within the hospitals and outside.
Finally, improved knowledge of molecular mechanisms controlling MDR should facilitate the development of novel therapies and will help develop deeper understanding of the microbial organism’s pathobiology. With MDR bacteria on the rise, new drugs have to be developed to combat bacterial antibiotic resistance, such as innate defense regulators, reactive oxygen species and microbial volatile compounds. In addition, searching for new ways to treat bacterial infections such as bacteriophages, their effective enzymes against bacteria and medicinal plants, could help in competing this MDR threat.
REFERENCES
Arnesen, S., Fagerlund, L. P. A. and Granum, P. E. (2008). From soil to gut: Bacillus cereus and its food poisoning toxins. FEMS Microbiology Reviews, (32): 579.606.
Boneca, I. G. and Chiosis, G. (2003). Vancomycin Resistance: Occurrence, Mechanisms And Strategies To Combat It. Expert Opinion On Therapeutic Targets, (7): 311-328.
Borer, A., Gilad, J., Yagupsky, P., Peled, N., Porat, N., Trefler, R., Shprecher-Levy, H., Riesenberg, K., Shipman, M. and Schlaeffer, F.(2002).Community-Acquired Methicillin-Resistant Staphylococcus aureus In Institutionalized Adults With Developmental Disabilities. Emerging Infectious Diseases, (8): 966-70.
Čižman, M. (2003). The use and resistance to antibiotics in the community. International journal of antimicrobial agents, (21): 297-307.
Ferreira, A. M., Bonesso, M. F., Mondelli, A. L., Camargo, C. H. and Cunha, M. D. L. R. (2013). Oxacillin Resistance and Antimicrobial Susceptibility Profile of Staphylococcus saprophyticus and Other Staphylococci Isolated from Patients with Urinary Tract Infection. Chemotherapy, (58) 482-491.
Jiang, S., Zeng, J., Zhou, X., and Li, Y. (2018). Drug Resistance and Gene Transfer Mechanisms in Respiratory/Oral Bacteria. Journal of Dental Research 97:002203451878265.
Kapoor, G., Saigal, S. and Elongavan, A. (2017). Action and resistance mechanisms of antibiotics: A guide for clinicians. J. Anaesthesiol. Clin. Pharmacol., (33):300-305.
Koksal, F., Yasar, H. and Samasti, M. (2009). Antibiotic resistance patterns of coagulase-negative staphylococcus strains isolated from blood cultures of septicemic patients in Turkey. Microbiological research, (164): 404-410.
Leach, K. L., Swaney, S. M., Colca, J. R., Mcdonald, W. G., Blinn, J. R., Thomasco, L. M., Gadwood, R. C., Shinabarger, D., Xiong, L. and Mankin, A. S. (2007). The site of action of oxazolidinone antibiotics in living bacteria and in human mitochondria. Molecular cell, (26): 393-402.
Maurya, N. and N. R. Agarwal (2019). A systematic short report on antibiotic resistance trends among bacteria isolated from sewage water. RJLBPCS, 5(1):33.
Raz, R., Colodner, R., Kunin, C. M. (2005). Who are you-Staphylococcus saprophyticus? CID (40): 896-898.
Roberts, M. C. )2005(. Update on acquired tetracycline resistance genes. FEMS microbiology letters, (245): 195-203.
Songer, J.G. (2010). Clostridia as agents of zoonotic disease. Veterinary Microbiology (140), 399-404.
Stark, L., Olofsson, M., Löfgren, S., Mölstad, S., Lindgren, P. and Matussek, A. (2013). Prevalence and molecular epidemiology of Staphylococcus aureus in Swedish nursing homes – as revealed in the SHADES study. Epidemiology and Infection. (142): 1310-1316.
Tanwar, J., Das, S., Fatima, Z. and Hameed, S. (2014). Multidrug resistance: an emerging crisis. Interdisciplinary perspectives on infectious diseases, 2014.
Tenover, F. C. (2006). Mechanisms of antimicrobial resistance in bacteria. The American journal of medicine, (119): S3-S10.
Turutoglu, H., Ercelik, S. and Ozturk, D. (2006). Antibiotic resistance of Staphylococcus aureus and coagulase-negative staphylococci isolated from bovine mastitis. Bulletin-Veterinary Institute In Pulawy, (50): 41.
Lebreton, F., Willems, R. J. L., and Gilmore, M. S. (2014). Enterococcus Diversity, Origins in Nature, and Gut Colonization.. In: Gilmore M. S., Clewell D. B., Ike Y., et al., editors. Enterococci: From Commensals to Leading Causes of Drug Resistant Infection [Internet]. Boston: Massachusetts Eye and Ear Infirmary
Livermore, D. M. (2003). Bacterial resistance: origins, epidemiology, and impact. Clinical infectious diseases, (36): S11-S23.
Mcdougal, L. K. and Thornsberry, C. (1986). The role of beta-lactamase in staphylococcal resistance to penicillinase-resistant penicillins and cephalosporins. Journal of Clinical Microbiology, (23):832-839.
McLauchlin J., Rees C.E. (2009). Genus Listeria. In: Whitman W.B. (ed.): Bergey’s Manual of Systematic Bacteriology, Volume 3. Springer-Verlag, New York, USA, 244-257.
Mengeloğlu, F. Z., Çakır, D. and Terzi, H. A. (2011). Comparison of resistance in isolates of Enterococcus faecalis and Enterococcus faecium. Journal of Microbiology and Infectious Diseases, (1):10-13.
Palazzo, I., Araujo, M. and Darini, A. (2005). First report of vancomycin-resistant staphylococci isolated from healthy carriers in Brazil. Journal of clinical microbiology, (43): 179-185.
Poole, K. (2002). Mechanisms of bacterial biocide and antibiotic resistance. Journal of Applied Microbiology, 92.
Von Wintersdorff, C. J. H., Penders, J., van Niekerk J. M., Mills N. D. , Majumder, S., van Alphen L. B. , Savelkoul, P. H. M. and Wolffs, P. F. G. (2016). Dissemination of Antimicrobial Resistance in Microbial Ecosystems through Horizontal Gene Transfer. Front Microbiol. (7): 173
Whitman, W. B., Coleman, D. C. and Wiebe, W. J. (1998). Prokaryotes: the unseen majority. Proceedings of the National Academy of Sciences, (95): 6578-6583.