1Department of Microbiology, Saveetha Dental College and Hospitals, Saveetha Institute of
Medical and Technical Sciences, Saveetha University, Chennai, Tamil Nadu, India.
Corresponding author email: smilinejames25@gmail.com
Article Publishing History
Received: 15/09/2021
Accepted After Revision: 25/12/2021
The epsA associated biofilm formation attributes to potent virulence in the drug resistant strains of Acinetobacter baumannii. This study is aimed to molecularly characterize the epsA gene among the multidrug resistant clinical isolates of A. baumannii and to assess the frequency of the same in different drug resistant groups. To detect the biofilm formation among the selected MDR strains of A. baumannii, semi-quantitative adherent bioassay was performed using crystal violet staining method. Further PCR amplification was done to screen the presence of epsA gene with further sequencing of the amplicons. Pearson’s correlation analysis was done to check the correlation of the occurrence of epsA gene with drug resistant strains (p-value<0.05). 58.9%, 31.5% and 0.9% of the strains were recorded as high grade, low grade and negative biofilm formers under biofilm assay.
The epsA gene was observed in 14 MDR strains (19.17%) of A. baumannii with an amplicon size of 451bp. Co-occurrence of epsA gene was 100% in β-lactam, cephem and folate resistant strains followed by 71.4% among aminoglycosides, 57.1% against carbapenems and 14.2% in fluoroquinolone and efflux pump mediated resistant strains. The findings of the study suggest the co-occurrence of epsA gene mediated biofilm formation among the multidrug resistant strains of A. baumannii. Further studies on the same helps in designing new vaccines and drugs for the prevention and treatment of A. baumannii infections.
A. Baumannii, Biofilm, Epsa, Multidrug Resistance, Virulence
Sarojini K, Girija A. S. S, Priyadharsini J.V. Molecular Detection of epsA-Mediated and Extracellular Polysaccharide-Mediated Biofilm Formation Among Multidrug Resistant Strains of Acinetobacter baumannii. Biosc.Biotech.Res.Comm. 2021;14(4).
Sarojini K, Girija A. S. S, Priyadharsini J. V. Molecular Detection of epsA-Mediated and Extracellular Polysaccharide-Mediated Biofilm Formation Among Multidrug Resistant Strains of Acinetobacter baumannii. Biosc.Biotech.Res.Comm. 2021;14(4). Available from: <a href=”https://bit.ly/3lgak7A“>https://bit.ly/3lgak7A</a>
Copyright © Sarojini et al., This is an open access article distributed under the terms of the Creative Commons Attribution License (CC-BY) https://creativecommns.org/licenses/by/4.0/, which permits unrestricted use distribution and reproduction in any medium, provide the original author and source are credited.
INTRODUCTION
Acinetobacter baumannii is a ubiquitous gram-negative and non-fermentative bacillus which causes a variety of diseases in humans. It exists mainly in health care settings such as hospitals and is recognized as one of the six dreadful nosocomial pathogens by the World Health Organization. Severe systemic complications linked with A. baumannii among hospitalized patients in intensive care units are associated with various infections like urinary tract infections, pneumonia, endocarditis, post-operative wound infections and septicemia have been recognized globally (Mayers et al. 2017; Matar 2018). The ability of A. baumannii to survive in the hospital setting is attributed with its biofilm formation. Initiation and formation of biofilms contributes to virulence in A. baumannii, as they were recognized as bacterial communities which enclosed in a matrix of extracellular material like DNA, proteins and polysaccharides (Ketter 2015; (Ramírez et al. 2021) .
Additionally, the biofilm of A. baumannii promotes the survival rate in both biotic and abiotic surfaces through its cell adhesiveness (Biswas and Rather 2019; González et al. 2021). A. baumannii is grouped as one of the ESKAPE organisms that is Enterococcus faecium, Staphylococcus aureus, Klebsiella pneumoniae, A. baumannii, Pseudomonas aeruginosa, and Enterobacter spp., that pose a global threat to human health and a therapeutic challenge to researchers due to the constant emerging of increasing resistance to drugs (Kyriakidis et al. 2021).
- baumannii infections account for >2% of all healthcare-associated infections in the Asia and the Middle East of which ~45% of all isolates are considered to be MDR, a rate up to four times increased when described in relation to other Gram-negative pathogenic organisms, such as P. aeruginosa and K. pneumoniae (Harding et al. 2018). Many phenotypes and genotypes vary in the expression of biofilm initiation, progression or development and distortion attributing to the virulence in A. baumannii. This is due to the different forms of the gene operons associated with the biofilm formation such as ompA, csu, csg, ptk, bap, epsA, kpsmII etc (Ahimou et al. 2007; Costerton et al. 2014). Amidst many genes, it is a known fact that epsA gene encodes for extracellular polysaccharide biofilms in A. baumannii. epsA based biofilms were also detected in many other bacteria resulting in the aggregation of the bacterial cells (Costerton et al. 2014; Chakravarty 2020).
These polysaccharide lines on the surface of A. baumannii organize into moderate to strong biofilms rendering effective protection for the bacterium to survive in harsh environments (Ghasemi et al. 2018). A recent study has documented its 95% occurrence among the drug resistant strains of A.baumannii (Zeighami et al. 2019). As an outcome of these statistics, the infections induced by carbapenem-resistant A. baumannii has been categorised in the critical group of all bacteria as a global threat to human health by WHO for prioritizing research and evolution of new antimicrobial treatments (Vázquez-López et al. 2020).
With this background, assessment on the correlation of the occurrence of epsA gene among the multi-drug resistant clinical isolates of A. baumannii would be a timely investigation as it is not so vivid in many studies from South India. Not much studies are documented related to the epsA based biofilm formation in the strains of hospitalized patients. This study is thus aimed to molecularly characterize epsA gene among the clinical isolates of A. baumannii with further comparative genomic assessments of the sequenced amplicons of the epsA gene (Vranceanu et al. 2020).
MATERIAL AND METHODS
The formation of the biofilms by the drug-resistant strains was investigated by culturing the cells in 96-well flat-bottomed microtiter plates as described earlier (Kouidhi et al. 2010). The detection of biofilm formation by semi-quantitative adherence assay was carried out in triplicates for each strain, with 200µl of the fresh broth culture in trypticase soy broth (HiMedia, Mumbai, India) with 0.25% glucose (w/v). The plate was incubated at 37°C /24 hrs with negative control (broth + 0.25% glucose) and positive control (known biofilm forming strain of A. baumannii earlier detected).
After incubation the wells were washed thrice with phosphate buffered saline (PBS), to remove the free cells, and the adhered bacteria were fixed using 95% ethanol/5 min and the plates were dried. Finally, all the wells were stained with 100µl of 1% w/v crystal violet solution (HiMedia), for 5 mins. Excess stains were removed by washing with distilled water and the wells were dried. Optical density was measured in the plate reader at 570 nm (OD570) and the biofilm formation was graded as high (OD570 ≥ 1), low (0.1 ≤ OD570 < 1) or negative (OD570 < 0.1) (Avila-Novoa et al. 2019).
73 different groups of drug-resistant strains of A. baumannii isolated in our earlier studies were maintained at -80⁰C in 80% / 20% (v/v) glycerol in LB medium in our repertoire, and were retrieved by incubating at 37⁰C for 24 hrs. Chromosomal DNA was extracted using the Qiagen DNA extraction kit in accordance with the manufacturer’s instructions (Inchai et al. 2015). The extracted genomic DNA was stored in -20⁰C until further use.
For the amplification of epsA, the PCR reaction mixture [15 µl] was prepared by adding 7.8 µl of 2x master mix [Taraka, Japan] in 5.6 µl of double distilled water with 0.31 µl of 100 pmol/ml concentration of the specific Primer and Primer [Eurofins Genomic India Pvt Ltd, Bangalore] of epsA genes. To the master mix 1 µl of the DNA was added and the amplification was carried out with PCR condition of 55°C as annealing temperature for 35 cycles in Eppendorf thermocycler, Germany.
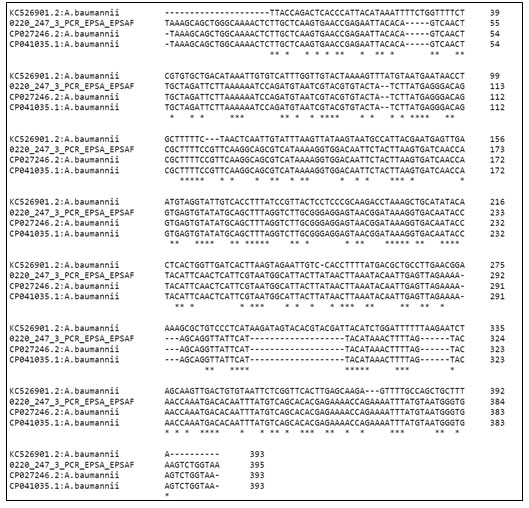
Using Big-Dye terminator cycle sequencing kit and 3730XL Genetic Analyzer the amplicon product of epsA were bidirectional sequenced. The obtained sequences of F and R primers we’re aligned using Bioedit Sequence Alignment Editor v7.2.5 which were subjected to BLAST (Basic Local Alignment Search Tool) for nucleotide similarity search. With the help of ClustalW software version 1.83 for DNA multiple sequence alignment using default parameters the sequences were aligned. For analyzing epsA sequences of A. baumannii strain WCHAB005078 (CP027246.2), A. baumannii strain KC526901.2 and A. baumannii strain 11W359501 (CP041035.1) were used as templates.
RESULTS AND DISCUSSION
Correlation of epsA with multidrug resistance; Semi quantitative adherent bioassay for biofilm formation showed 58.9% (43/73) under high grade, 31.5% (23/73) under low grade and 0.9% (7/73) to be negative. Amidst the 43 strains of high-grade biofilm formers all were multidrug resistant (100%; 43/43) exhibiting resistance against more than 3 classes of the antibiotics tested followed by 91.3% (21/23) under low grade biofilm formers. Under the negative biofilm formers, only one strain was drug resistant. Pearson correlation analysis yielded positive value suggesting the correlation of the occurrence of epsA gene with drug resistant strains (p-value<0.05).
From the screened 73 genomes of multi-drug resistant strains of A. baumannii, 19.17% (n=14) showed positive amplicons for the epsA gene associated with biofilm formation (Figure 1). Correlation of epsA occurrence was high (100%; 14/14) in the resistant groups of beta-lactam inhibitors (piperacillin and tazobactam), cephalosporins (ceftazidime, cefepime, ceftriaxone, ceftriaxime) and folate drugs (co-trimoxazole).
It was followed by 71.4% (n=10/14) aminoglycosides (amikacin and kanamycin) and 57.1% (8/14) occurrence among the strains resistant to carbapenems (doripenem, meropenem and imipenem). The least occurrence was observed with 14.2% (2/14) among the strains resistant to fluoroquinolones (ciprofloxacin and levofloxacin) and efflux pumps (tetracycline, doxycycline and minocycline) drugs (Figure 2). None of the control susceptible strains have shown positive for epsA gene. Figure 3 and 4 show the sequence chromatogram and the multiple sequence alignment of the epsA amplicon.
Table 1: Primer sequence and PCR conditions to detect epsA gene in multi-drug resistant strains of A. baumannii
| Gene of target | Primer details | Annealing temp | Amplicon size |
| epsA | AGCAAGTGGTTATCCAATCG
ACCAGACTCACCCATTACAT |
55
|
451 bp |
Figure 1: Electropherogram of epsA gene product of size 451bp in lane 1 and 2 with marker ladder of size 100bp (M)
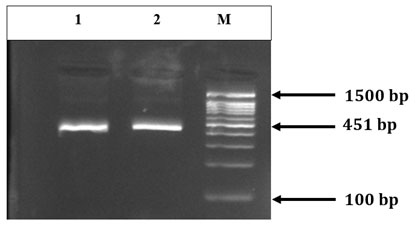
Figure 2: Frequency of epsA gene among different groups of antibiotic resistant strains of A. baumannii
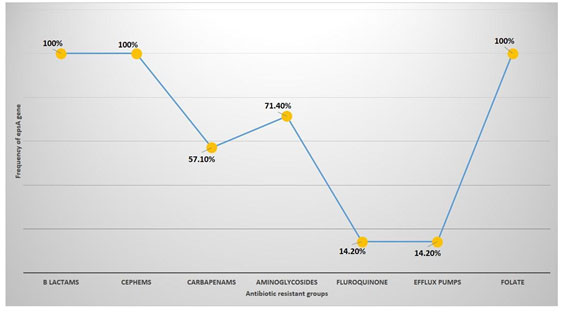
Figure 3: Partial sequence chromatogram of epsA gene from the amplicon of A. baumannii
Figure 4: Partial sequence alignment of epsA gene from the present study (Ab) with reference sequences available in the database. The deleted regions are depicted as dashes (–), mismatch as gap ( ) and conserved sequences as star (*).
In recent years, A. baumannii is considered as a potent nosocomial pathogen and in the past decade there has been a high prevalence of multidrug resistant clinical isolates in hospitalized patients, ranging from 21–95% (Zhang et al. 2016).
Assessment on its virulence properties in harsh environmental niches is highly attributed to its ability of biofilm formation. Biofilm associated bacteria is generally associated with two properties namely, the increased synthesis of exopolysaccharides and the development of antibiotic resistance. Increased production of these extracellular polysaccharides (EPS) in A. baumannii produces a protective environment causing difficulty in antibiotic penetration leading to the development of resistance (Lewis 2001; Høiby et al. 2010)
The spread of antibiotic resistance is by the ability of the bacterial pathogen to transfer genes horizontally and is commonly associated with biofilm formers (Ghasemi et al. 2018). Amidst various biofilm associated genes. epsA is considered as a potent contributor for its correlation with drug resistance as the exopolysaccharide layers can influence the entry of antibiotics (Donlan and Costerton 2002; Leclercq et al. 2013; Tchuinte et al. 2019; Bavelaar et al. 2021).
Thus, the present investigation is undertaken to molecularly assess the same and its correlation of its occurrence among the multi-drug resistant strains in hospital environments. A strong positive correlation of epsA and biofilm formation had already been documented, and in various other studies its contribution ranged from 30.2% respectively ( Russo et al. 2010). The role of epsA in the biofilm formation on abiotic surfaces also reveal the existence of A. baumannii amidst various disinfectants and antimicrobial agents aiding in the transfer of resistance genes with the participating strains too (Sung 2018).
Many studies reporting the confirmation of its association with the biofilm formation in nosocomial environments, the biofilm formation assay performed in the present study with the routine crystal violet staining method graded the strains as high- and low-grade biofilm formers. The high frequency of epsA observed in the present study hypothesizes that it would have aided A. baumannii to survive and persist in the hospital habitats, as the gene was detected by PCR from the clinical isolates of the same. This data is again a key fact that this sort of identification of the A. baumannii virulence factors periodically would offer more effective ways to eradicate them from the biotic and abiotic surfaces of the hospitals (Kongthai et al. 2021).
Our reports correlate with similar earlier studies associated with the presence of epsA genes in drug resistant clinical isolates of A. baumannii. Highest frequency of 95% epsA gene among MDR strains of A. baumannii was reported from Iran. In a similar study by Assaad et al. (2021) had documented 60% of the multi-drug resistant isolates of A. baumannii showing positivity for epsA (Zeighami et al. 2019; Asaad et al. 2021). In a study by Kongthai et al. (2021) 30.2% of the biofilm producers were epsA positive and were also multidrug resistant (Kongthai et al. 2021).
In the same study, 86.2% epsA gene positive strains were multi drug resistant and the resistance ranged from 50% to 80% against different classes of antibiotics viz., piperacillin, cefixime, ciprofloxacin, levofloxacin, ceftazidime, gentamicin, ticarcillin and imipenem. In a study by Safari et al. (2015) 38% of the isolates encoded epsA gene with strong biofilms (Safari et al. 2015). In a similar study conducted by Sung et al 30% of the multi-drug resistant clinical isolates of A. baumannii showed the presence of epsA (Sung 2018). The present study also documents the frequency of epsA gene in different groups of antibiotics routinely employed against A. baumannii in hospital set-ups globally (Pompilio et al. 2021).
Results of the study showed high correlation of epsA with beta lactam, cephems and folate resistant isolates, and may be related to the presence of epsA gene in all the resistant isolates. This is higher when compared to the study by Badave et al. (2015) where the epsA positive strains were 55.5% resistance to ampicillin and 42.6% resistance to piperacillin (beta lactams), 52.08% were resistant to ceftazidime, and 50% were resistant to ciprofloxacin. In contrast, a study in South India, biofilm positive Acinetobacter showed resistance to both ceftazidime (95%) and cefepime (95%) (Badave 2015; Zeighami et al. 2019; Asaad et al. 2021).
The same study also had documented epsA positive biofilm formers showing 85% resistance to ciprofloxacin. Carbapenem resistance in A. baumannii is based on the production of carbapenemases or synergistic effects between carbapenemases, porin modifications or loss or modification of the penicillin-binding proteins (Towner et al. 1991; Poirel and Nordmann 2006). In our study, epsA mediated biofilm formers showed on 57.1% resistance to carbapenem group of drugs which is correlating with the study conducted by Zeighami et al.
(2019), where 100% of isolates showed resistance to imipenem and this result is consistent with observations reported from various parts of the world which explain the high risk of failure of carbapenem treatment in A. baumannii infections but yet another study showed only 38.4% were resistant to imipenem (Rahbar et al. 2010; Peerayeh and Karmostaji 2015; Badave 2015; Saffari et al. 2017).
Overexpression of efflux pumps had been postulated to be a first step towards the development of a MDR phenotype in bacteria and most of them lead to the resistance to tetracyclines. A strong association of epsA genes was observed in the present study with 14.2% of the strains showing resistance against the cycline group of drugs. The present study observed the correlation of epsA genes with 100% resistant strains of drugs involved with folate pathway with the lowest occurrence of 14.2% among the strains resistant to fluoroquinolones. Similarly, correlation of epsA occurrence (71.4%) in aminoglycoside resistant strains of A. baumannii, correlates with the earlier study with 70% frequency and higher when compared to 53.4% amikacin resistant isolates in an another study (Bambeke et al. 2000; Piddock 2006; Kongthai et al. 2021).
CONCLUSION
The findings of the present study investigated the presence of biofilm-forming epsA gene among the clinical isolates of A. baumannii. In spite of the effective infection management, it’s thus alarming to note the augmented prevalence of biofilm-associated epsA gene among the multidrug-resistant clinical isolates of A. baumannii. Thus, it would be timely to suggest the periodical monitoring of the hospitals and laboratories for the presence of epsA mediated biofilm producing resistant groups of A. baumannii. This would also aid in designing new vaccines and drugs for the prevention and treatment of A. baumannii infections.
Conflict of interests Authors declare no conflict of interest to disclose.
REFERENCES
Ahimou, F., Semmens, M.J., Haugstad, G. et al. (2007). Effect of protein, polysaccharide, and oxygen concentration profiles on biofilm cohesiveness. Applied and environmental microbiology, 73(9), pp.2905-2910
Asaad, A.M., Ansari, S., Ajlan, S.E. et al. (2021). Epidemiology of Biofilm Producing Acinetobacter baumannii Nosocomial Isolates from a Tertiary Care Hospital in Egypt: A Cross-Sectional Study. Infection and drug resistance, 14, p.709.
Avila-Novoa, M.G., Solís-Velázquez, O.A., Rangel-Lopez, D.E., et al. (2019). Biofilm formation and detection of fluoroquinolone-and carbapenem-resistant genes in multidrug-resistant Acinetobacter baumannii. Canadian Journal of Infectious Diseases and Medical Microbiology, 2019.
Badave, G.K. and Kulkarni, D., (2015). Biofilm producing multidrug resistant Acinetobacter baumannii: an emerging challenge. Journal of clinical and diagnostic research: JCDR, 9(1), p.DC08.
Bavelaar, H., Justesen, U.S., Morris, T.E., et al. (2021). Development of a EUCAST disk diffusion method for the susceptibility testing of rapidly growing anaerobic bacteria using Fastidious Anaerobe Agar (FAA): a development study using Bacteroides species. Clinical Microbiology and Infection, 1198-743.
Biswas, I. and Rather, P.N., (2019). Acinetobacter baumannii. Springer, 1-337.
Chakravarty, B., (2020). Genetic mechanisms of antibiotic resistance and virulence in Acinetobacter baumannii: background, challenges and future prospects. Molecular biology reports, 47(5), pp.4037-4046.
Costerton, J.W. and Stewart, P.S., (2000). Biofilms and device‐related infections. Persistent bacterial infections, pp.423-439.
Donlan, R.M. and Costerton, J.W., (2002). Biofilms: survival mechanisms of clinically relevant microorganisms. Clinical microbiology reviews, 15(2), pp.167-193.
Ghasemi, E., Ghalavand, Z., Goudarzi, H., et al. (2018). Phenotypic and genotypic investigation of biofilm formation in clinical and environmental isolates of Acinetobacter baumannii. Archives of Clinical Infectious Diseases, 13(4).
Harding, C.M., Hennon, S.W. and Feldman, M.F., (2018). Uncovering the mechanisms of Acinetobacter baumannii virulence. Nature Reviews Microbiology, 16(2), pp.91-102.
Høiby, N., Bjarnsholt, T., Givskov, M., et al. (2010). Antibiotic resistance of bacterial biofilms. International journal of antimicrobial agents, 35(4), pp.322-332.
Inchai, J., Liwsrisakun, C., Theerakittikul, T., et al. (2015). Risk factors of multidrug-resistant, extensively drug-resistant and pandrug-resistant Acinetobacter baumannii ventilator-associated pneumonia in a Medical Intensive Care Unit of University Hospital in Thailand. Journal of Infection and Chemotherapy, 21(8), pp.570-574.
Ketter, P.M., (2015). Study of virulence factors associated with Acinetobacter baumannii systemic and gastrointestinal infections. The University of Texas at San Antonio.
Kongthai, P., Thummeepak, R., Leungtongkam, U., et al. (2021). Insight into Molecular Epidemiology, Antimicrobial Resistance, and Virulence Genes of Extensively Drug-Resistant Acinetobacter baumannii in Thailand. Microbial Drug Resistance, 27(3), pp.350-359.
Kyriakidis, I., Vasileiou, E., Pana, Z.D. et al. (2021). Acinetobacter baumannii Antibiotic Resistance Mechanisms. Pathogens, 10(3), p.373.
Leclercq, R., Cantón, R., Brown, D.F., et al. (2013). EUCAST expert rules in antimicrobial susceptibility testing. Clinical Microbiology and Infection, 19(2), pp.141-160.
Lewis, K., (2001). Riddle of biofilm resistance. Antimicrobial agents and chemotherapy, 45(4), pp.999-1007.
Matar, G.M., (2018). Pseudomonas and Acinetobacter: From drug resistance to pathogenesis. Frontiers in cellular and infection microbiology, 8, p.68.
Mayers, D.L., Sobel, J.D., Ouellette, M., et al. (2017). Antimicrobial Drug Resistance: Clinical and Epidemiological Aspects, Springer, 2.
Peerayeh, S.N. and Karmostaji, A., (2015). Molecular identification of resistance determinants, integrons and genetic relatedness of extensively drug resistant Acinetobacter baumannii isolated from hospitals in Tehran, Iran. Jundishapur journal of microbiology, 8(7).
Piddock, L.J., (2006). Clinically relevant chromosomally encoded multidrug resistance efflux pumps in bacteria. Clinical microbiology reviews, 19(2), pp.382-402
Poirel, L. and Nordmann, P., (2006). Carbapenem resistance in Acinetobacter baumannii: mechanisms and epidemiology. Clinical Microbiology and Infection, 12(9), pp.826-836.
Pompilio, A., Scribano, D., Sarshar, M., et al. (2021). Gram-Negative Bacteria Holding Together in a Biofilm: The Acinetobacter baumannii Way. Microorganisms, 9(7), p.1353.
Prieto, L., Villaseñor, B. and Martínez, N.M., (2021). Carbapenamase OXA-23 like-producing Acinetobacter baumanii epidemic outbreak in a hospitalization unit. Revista Espanola de Salud Publica, 95.
Rahbar, M., Mehrgan, H. and Aliakbari, N.H., (2010). Prevalence of antibiotic-resistant Acinetobacter baumannii in a 1000-bed tertiary care hospital in Tehran, Iran. Indian Journal of Pathology and Microbiology, 53(2), p.290.
Ramírez, S.C., Hamidian, M., Zarrilli, R. et al. (2021). ORIGINAL RESEARCH published: 17 June 2020. Genomic Basis of Antibiotic Resistance and Virulence in Acinetobacter, p.1176661.
Russo, T.A., Luke, N.R., Beanan, J.M., et al. (2010). The K1 capsular polysaccharide of Acinetobacter baumannii strain 307-0294 is a major virulence factor. Infection and immunity, 78(9), pp.3993-4000.
Safari, M., Nejad, A.S.M., Bahador, A., et al. (2015). Prevalence of ESBL and MBL encoding genes in Acinetobacter baumannii strains isolated from patients of intensive care units (ICU). Saudi journal of biological sciences, 22(4), pp.424-429.
Saffari, F., Monsen, T., Karmostaji, A., et al. (2017). Significant spread of extensively drug-resistant Acinetobacter baumannii genotypes of clonal complex 92 among intensive care unit patients in a university hospital in southern Iran. Journal of medical microbiology, 66(11), pp.1656-1662.
Sung, J.Y., (2018). Molecular characterization and antimicrobial susceptibility of biofilm-forming Acinetobacter baumannii clinical isolates from Daejeon, Korea. Korean Journal of Clinical Laboratory Science, 50(2), pp.100-109.
Tchuinte, P.L.S., Rabenandrasana, M.A.N., Kowalewicz, C., et al. (2019). Phenotypic and molecular characterisations of carbapenem-resistant Acinetobacter baumannii strains isolated in Madagascar. Antimicrobial Resistance & Infection Control, 8(1), pp.1-9.
Towner, K.J., Bergogne-Bérézin, E. and Fewson, C.A., (2013). The biology of acinetobacter: Taxonomy, clinical importance, molecular biology, physiology, industrial relevance, Springer Science & Business Media, 57.
Van Bambeke, F., Balzi, E. and Tulkens, P.M., (2000). Antibiotic efflux pumps. Biochemical pharmacology, 60(4), pp.457-470.
Vázquez-López, R., Solano-Gálvez, S.G., Juárez Vignon-Whaley, J.J., et al. (2020). Acinetobacter baumannii resistance: a real challenge for clinicians. Antibiotics, 9(4), p.205.
Vrancianu, C.O., Gheorghe, I., Czobor, I.B. et al. (2020). Antibiotic resistance profiles, molecular mechanisms and innovative treatment strategies of Acinetobacter baumannii. Microorganisms, 8(6), p.935.
Zeighami, H., Valadkhani, F., Shapouri, R., et al. (2019). Virulence characteristics of multidrug resistant biofilm forming Acinetobacter baumannii isolated from intensive care unit patients. BMC infectious diseases, 19(1), pp.1-9.
Zhang, D., Xia, J., Xu, Y., et al. (2016). Biological features of biofilm-forming ability of Acinetobacter baumannii strains derived from 121 elderly patients with hospital-acquired pneumonia. Clinical and experimental medicine, 16(1), pp.73-80.


