1Department of Plant Molecular Biology and Biotechnology, C. P. College of Agriculture (CPCA), Sardarkrushinagar Dantiwada Agricultural University (SDAU), Sardarkrushinagar-385506, Gujarat, India
2Shree P. M. Patel Institute of Integrated M. Sc. in Biotechnology, Anand-388001, Gujarat, India
3Department of Biotechnology, Mehsana Urban Institute of Sciences, Ganpat University, Ganpat Vidyanagar–384012, Gujarat, India
4College of Basic Science and Humanities, SDAU, Sardarkrushinagar-385506, Gujarat, India
5Centre of Seed Technology, S. D. Agricultural University, Sardarkrushinagar-385506, Gujarat, India
Corresponding author Email: darshanbiotech1@gmail.com
Article Publishing History
Received: 24/12/2018
Accepted After Revision: 01/03/2019
Potato (Solanum tuberosum L.) is the fourth most important food crop in the world and an important vegetable crop. The genus Solanum consists of 220 tuber containing species of which seven tuber-bearing species is used for commercial cultivation. Potato is a self-pollinated crop with cross-pollination up to 2.54 percent. Use of molecular markers to determine genetic variation, genetic diversity and evolutionary relatedness is becoming more popular for the assessment of diversity among cultivars of crop species of from various geographical origins. In the present study, total 42 RAPD markers have been employed, out of which 21 primers were polymorphic with 66.95% average polymorphism and 0.783 average PIC value, indicating higher informativeness of primers. In the present study, total 25 SSR markers have been employed out of these only 4 potent SSR primers were polymorphic. For SSR markers, average polymorphism was 58.33% and mean PIC value was 0.712. The results of current study indicate that RAPD and SSR markers used in the study have very promising polymorphism and PIC values hence, seemed to be good for the molecular characterization and assessing genetic relationship among genotypes of potato.
Potato (Solanum Tuberosum L.), Molecular Markers, Microsatellite, Rapd, Ssr
Kapuria M, Dharajiya D, Pachchigar K, Chauhan R. M. Molecular Characterization and Genetic Diversity of Indian Potato, (Solanum Tuberosum L.) Germplasms Using Microsatellite and RAPD Markers. Biosc.Biotech.Res.Comm. 2019;12 (1).
Kapuria M, Dharajiya D, Pachchigar K, Chauhan R. M. Molecular Characterization and Genetic Diversity of Indian Potato, (Solanum Tuberosum L.) Germplasms Using Microsatellite and RAPD Markers. Biosc.Biotech.Res.Comm. 2019;12(1). Available from: https://bit.ly/2HJDdps
Copyright © Kapuria et al., This is an open access article distributed under the terms of the Creative Commons Attribution License (CC-BY) https://creativecommns.org/licenses/by/4.0/, which permits unrestricted use distribution and reproduction in any medium, provide the original author and source are credited.
Introduction
Potato (Solanum tuberosum L.) originated from South America is one of the major vegetable crops of the world which is also the fourth most important food crop in the world, after maize, wheat, and rice (Ghebreslassie et al., 2016; Majeed and Muhammad, 2018). In India, it was introduced in early seventeenth century possibly by the Portuguese or by the Britishers (Pal and Nath, 1951; Kapuria et al., 2016). It is a source of low cost energy to the human diet hence, it is considered as an economy food (Pandey and Sarkar, 2005). Globally, the major potato growing nations are China, India, Russia, Ukraine and United States of America (Bradshaw, 2019). The genus Solanum consists of 220 tuber-containing species among which seven tuber bearing species exploited for commercial cultivation (Hawkes and Jackson, 1992). The most important feature in potato taxonomy is the variation in ploidy level. Potato is a self-pollinated crop and the level of cross-pollination is up to 2.54 percent (Kapuria et al., 2016; Wang et al., 2019).
Knowledge of the genetic diversity and relationships among the varieties/cultivars are very beneficial to recognize the gene pool, to recognize the gaps in germplasm collections and to improve effective conservation and management approaches. Use of DNA markers for the identification of genetic variation, genetic diversity and evolutionary relatedness can be useful in identifying genetic/population structure as well as diversity among cultivars from various geographical origins. Due to their simplicity, reliability and cost effectiveness, PCR based methods are in demand (Parita et al., 2018).
Different PCR based techniques have been developed during the last two decades, each with specific benefits and drawbacks. The random amplified polymorphic DNA (RAPD) marker system is rapid, simple and requires no prior sequence information in which one random 10-mer primer is used (Welsh and McClelland, 1990; Williams et al., 1990). The dominant nature of inheritance of RAPD is considered as its drawback. Among different classes of molecular markers, simple sequence repeats (SSR)/microsatellites which are tandem repeats of 1-6 nucleotide long DNA motifs gaining importance in plant breeding and genetics due to their co-dominant inheritance, multi-allelic nature, relative abundance, hyper variability, reproducibility, good genome coverage including organellar genomes, chromosome specific location, amenability to automation and high throughput genotyping (Oliveira et al., 2006; Walunjkar et al., 2013). The PCR based techniques have been used for DNA fingerprinting and genotyping in potato including RAPD (McGregor et al., 2000; Ghislain et al., 2006; Chimote et al., 2007; Rocha et al., 2010; Gorji et al., 2011; Onamu et al., 2016; Ayman et al., 2018), SSR (McGregor et al., 2000; Braun and Wenzel, 2004; Feingold et al., 2005; Ghislain et al., 2006; Chimote et al., 2007; Ghislain et al., 2009; Rocha et al., 2010; Favoretto et al., 2011; Maras et al., 2017; Ahmed et al., 2018; Tiwari et al., 2019), ISSR (McGregor et al., 2000; Bornet et al., 2002; Gorji et al., 2011; Vanishree et al., 2016) and AFLP (McGregor et al., 2000; Braun and Wenzel, 2004; Solano Solis et al., 2007; Jian et al., 2017). The present work was conducted to evaluate the phylogenetic relatedness of Indian potato genotypes using RAPD and SSR markers.
Materials and Methods
Plant material used in the study
Plant material consisted of young leaves sampled from young plants of 15 diverse Indian potato germplasms (Table 1). They were grown in Botanical Garden, C.P. College of Agriculture, SDAU, Sardarkrushinagar, Gujarat, India.
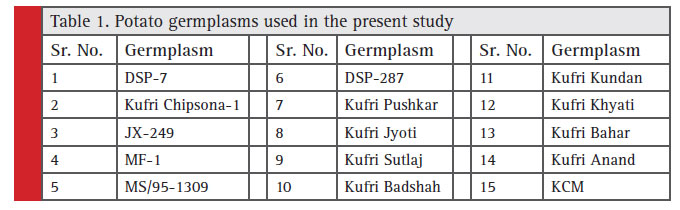 |
Table 1: Potato germplasms used in the present study |
Genomic DNA extraction and quantification
Genomic DNA was extracted from tender fresh leaves of each germplasm by using CTAB (Cetyl Trimethyl Ammonium Bromide) method described by Doyle and Doyle, (1990) with minor modifications. The quality and integrity of DNA were checked by electrophoresis using 0.8% agarose gel. The DNA samples were quantifi ed on UV spectrophotometer (BioSpectrometer, Eppendorf, Germany). The stocks were diluted to a fi nal concentration of 30 ng/μl of DNA and used for further work.
PCR amplifi cation
RAPD analysis
Forty two primers were selected and used to ascertain polymorphism among diverse germplasms of potato. The RAPD primers used in present study are UBC, OPA, OPB, OPC, OPH, OPG, OPW and OPX series and polymorphic primers used in the analysis are given in Table 2. PCR was carried out as method given by Yasmin et al., (2006) with minor modifications in a total reaction volume of 25 μl. The PCR mixture consisted of 1 X PCR buffer, 10 mM dNTPs, 2.0 mM MgCl2, 1 U of Taq DNA polymerase, 20 pmol/μl Primer and 30 ng/μl DNA template. All amplifications were carried out on a eppendorf thermal cycler using PCR conditions initial denaturation at 94ºC (4 min) one time, denaturation at 94 ºC for 1min, annealing at Tm of primer for 45 sec and extension at 72ºC for 2 min for 35 cycles with final extension of 72ºC for 7 min.
SSR analysis
PCR was carried out as method given by Ghebreslassie et al., (2016) with minor modifications in a total reaction volume of 25 μl. The PCR mixture consisted of 1 X PCR buffer, 10 mM dNTPs, 2.0 mM MgCl2, 1 U Taq DNA polymerase, 20 pmol/μl of primer pair and 30ng/ μl of template DNA. The polymorphic primers used in present study are given in Table 2. PCR conditions consisted of initial denaturation at 94ºC (4 min) for first cycle only, denaturation at 94 ºC for 30 sec, annealing at Tm of primer for 1 min and extension at 72ºC for 1 min for 35 cycles with fi nal extension of 72ºC for 6 min.
Resolution of amplified products
The amplifi ed products of RAPD and SSR were resolved on 1.5% and 3.0% agarose gel (Dharajiya et al., 2017). The gel was stained with ethidium bromide (10µl/100ml). The standard DNA marker (100 bp) was also run along with the samples. After electrophoresis, the gel was carefully taken out of the casting tray and photographed using AlphaEaseFC 4.0.0 Gel Documentation system (Alpha Innotech Corporation, USA).
Analysis of RAPD and SSR data
Data were scored for computer analysis on the basis of the presence (1) or absence (0) of the amplified DNA fragments. The data were entered into the binary matrix and subsequently analyzed using PAleontological STatistics (PAST) -Version 3.18 (Hammer et al., 2001) was used for genetic diversity evaluation. Coefficients of similarity were calculated by using Jaccard’s similarity co-effi cient (Jaccard, 1908) and cluster analysis was performed by using the Un-weighted Pair Group Method with Arithmetic Mean (UPGMA) function of PAST version 3.18. The Relationship between the potato genotypes was graphically represented in the form of dendrograms by using the cluster analysis function of the software. In this method the dendrogram and similarity matrix were correlated to find the goodness-of-fit of the dendrogram constructed based on the similarity coefficients. The marker data were then standardized for Principal Component Analysis (PCA). The software program AlphaEaseFC version 6.0.0 developed by Alpha Innotech Corporation, USA was used for determining the Molecular Weight (MW) of bands separated on the gel. The Polymorphism Information Content (PIC) value for each locus was calculated on the basis of allele frequency by the formula given by Anderson et al., (1993). The polymorphism percentage was calculated as per the method suggested by Smith et al., (1997).
Results and Discussion
Molecular marker analysis
Total 42 RAPD primers were used for screening of 15 potato germplasms. Out of 42 primers, 21 were polymorphic which amplifi ed a total of 116 reproducible DNA fragments. Out of total 116 DNA fragments, 79 fragments were polymorphic with the mean number of polymorphic bands per primer was 3.76. The size of amplified fragments varied from 140 to 2059 bp. The highest number of amplifi ed bands (8) was exhibited by primer UBC 31 and UBC 34 whereas lowest number of amplified bands (3) was produced by primer OPW 4. The highest polymorphism (100%) was exhibited by two primer OPA 8 and OPB 18, while the lowest polymorphism (20.00 %) was evinced with OPW 8. The average polymorphism detected by the RAPD loci in the present investigation was 66.95 % (Table 2).
Out of 25 SSR primers used in the study, 4 were polymorphic which amplifi ed a total of 18 reproducible fragments among which 11 fragments were polymorphic. The mean number of polymorphic bands was 2.75. The size of PCR amplifi ed fragments varied from 125 to 1899 bp. The highest number of amplifi ed band (6) was exhibited by primer STWIN12G and STCPKIN 3 and the lowest number of amplifi ed bands (2) was exhibited by primer STPRINPSG. The highest polymorphism (66.66) was exhibited by two primers viz., STWIN12G and STCPKIN 3, while the lowest polymorphism (50.00%) was evinced with STPRINPSG and STACCS 3. The average polymorphism detected by the SSR loci in the present investigation was 58.33 % (Table 2).
The distribution of the primers used in the study according to the PIC values and per cent polymorphism is given in the Fig. 1. Most of the RAPD primers (11) have PIC value from 0.7 to 0.8 whereas two SSR primers have PIC value from 0.8 to 0.9. Most of the RAPD primers have per cent polymorphism more than 60 %. It indicates that these primers possess good importance in the diversity analysis in potato.
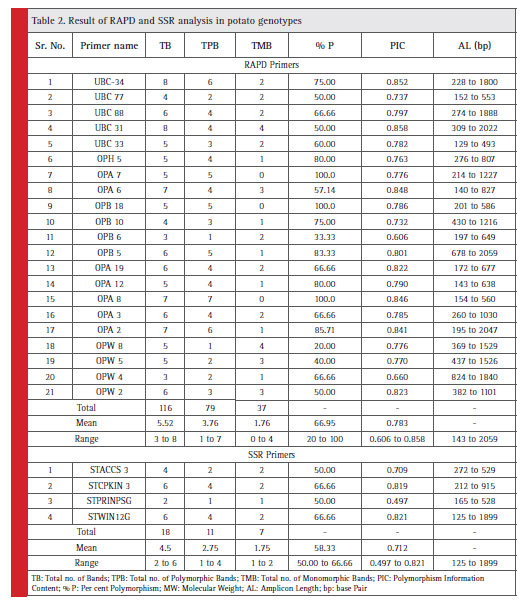 |
Table 2: Result of RAPD and SSR analysis in potato genotypes |
Construction of dendrogram
RAPD based dendrogram
Jaccard’s co-efficients were used to compare set of variables and to generate similarity matrix. Jaccard’s co-efficients for all genotypes as per RAPD analysis are shown in Table 3. Similarity indices were estimated on the basis of twenty one RAPD primers ranged from 0.56 (between DSP-287 and Kufri Chipsona-1) to 0.82 (between Kufri Kundan and Kufri Sutluj). UPGMA (unweighted pairgroup method with arithmetic mean) dendrogram was prepared by using Jaccard’s similarity co-effi cients (Fig 2. (A)). The dendrogram clustered with the data generated by all primers and their amplicons grouped the 15 genotypes into two major clusters i.e., Cluster A and Cluster B. Dendrogram showed two major clusters with co-efficient value 0.7564. The cluster A contained only one genotype, namely, Kufri Pushkar. The cluster B was further grouped in three clusters B1, B2 and B3. The
cluster B1 contained 11 genotypes. The cluster B2 and B3 contains two and one genotype, respectively.
SSR based dendrogram
Jaccard’s similarity co-effi cients (Table 4) were estimated on the basis of four SSR primers, ranged from 0.47 (Kufri Badshah and Kufri Kundan with DSP-287) to 1.0 (between Kufri Kundan and Kufri Badshah). UPGMA dendrogram was prepared by using Jaccard’s similarity co-efficients which was grouped into two main clusters i.e., Cluster A and Cluster B (Fig 2. (B)). Dendrogram showed two major clusters with co-efficient value 0.8306. The cluster A was further divided into two clusters A1 and A2. The cluster A1 contained 6 genotypes and cluster A2 contained 8 genotypes. The cluster B was dividing into one group containing only one genotype DSP-287. Genotypes Kufri Badshah and Kufri Kundan had highest similarity.
Principle Component analysis (PCA)
In the PCA plot, derived from the RAPD genotyping data, it can be observed that Kufri Pushkar and Kufri Sutluj is placed farthest from Kufri Anand in the 1st coordinate (X-axis), while Kufri Bahar and MF-1 were placed farthest in the 2nd coordinate (Y-axis) (Fig. 3 (A)). Most of the varieties with moderately resistances were located in 1st coordinate right side in the plot including MF-1, DSP-287 and MS/95-1309 except Kufri Sutlaj. In the PCA plot derived from the SSR genotyping data, it can be observed that DSP-287 and Kufri Sutluj is placed farthest from Kufri Anand in the 1st coordinate (X-axis), while Kufri Jyoti and DSP-287 were placed farthest in the 2nd coordinate (Y-axis) (Fig. 3 (B)). Varieties with moderately resistance located on the right side of 1st coordinate in the plot including the MF-1 and MS/95- 1309 except Kufri Sutluj and DSP-287.
In potato, various studies have been previously reported at molecular level deciphering variation across accession and varieties. Center wise studies also have been performed that indicating within the center diversity. Previously, RAPD analysis has been employed in many literature for checking molecular polymorphism and allelic variation McGregor et al., (2000), Yasmin et al., (2006), Chimote et al., (2007), Onamu et al., (2016) and Ayman et al., (2018). In the present study, total 42 RAPD markers have been employed out of which 50% of primers accounts for mean polymorphism greater than 66.95% with PIC 0.783, indicating higher informativeness of primers. Previously, Chimote et al., (2007), Gorji et al., (2011) and El Komy et al., (2012) found mean polymorphism about 20%, 31% and 57.4% which was less then present result, indicating more superior polymorphic potential of primers used in the present study and its employment for further research by other in future. Moreover, Gorji et al., (2011) found 0.28 mean PIC value which was very less as compared to the mean PIC value of RAPD (0.78) in the present research. Recently, Onamu et al., (2018) used 19 RAPD markers for the assessment of genetic diversity among 35 potato accessions and reported 81.45% polymorphism which was higher as compare to the current investigation.
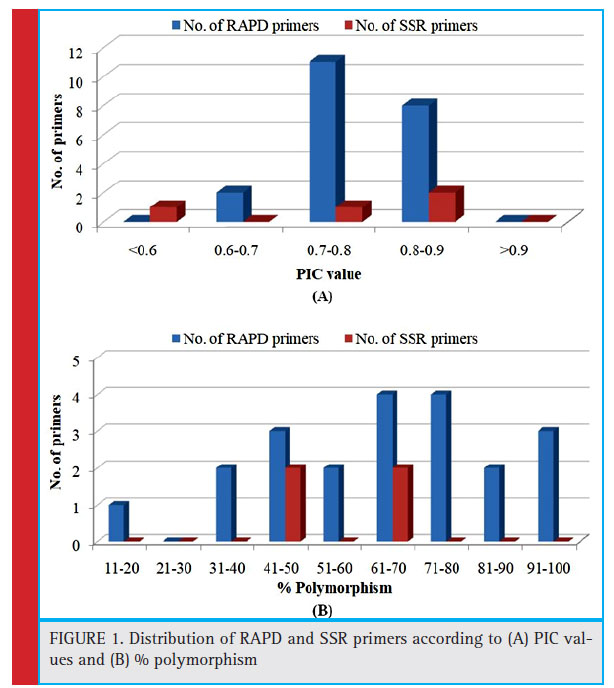 |
Figure 1: Distribution of RAPD and SSR primers according to (A) PIC values and (B) % polymorphism |
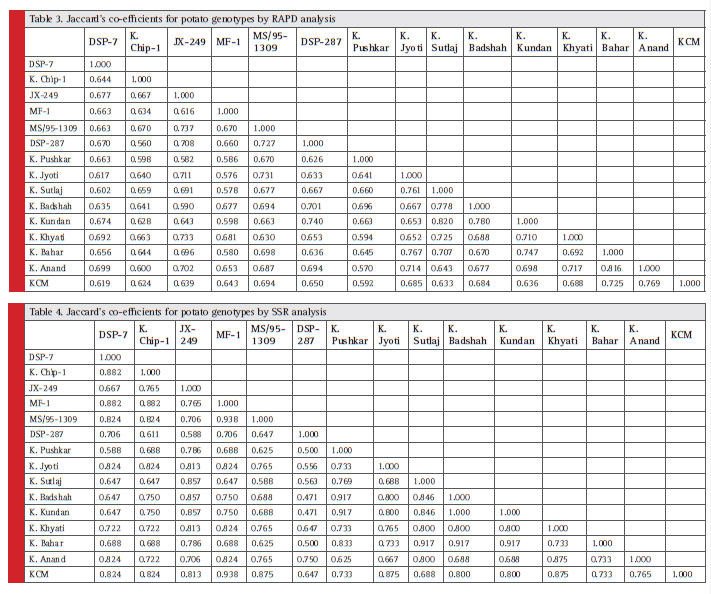 |
Table 3, 4 |
Among DNA markers, microsatellites have been chosen over RAPD because of their co-dominant behavior, multiallelism, reproducibility, and high level of polymorphism detected. In potato, few researchers such as Mc Gregor et al., (2000), Feingold et al., (2005), Ghislain et al., (2006), Chimote et al., (2007), Sharma and Nandineni, (2014), Maras et al., (2017), Ahmed et al., (2018) and Tiwari et al., (2019) have successfully used SSR markers. In the present study, total 25 SSR markers have been employed out of these only 4 potent SSR primers were polymorphic. Average polymorphism was 58.33% and mean PIC value was 0.712. Previously, Chimote et al., (2007) work with SSR marker for with general morphological character turns to only mean 19.5% polymorphism with almost same genotype indicating more superior polymorphic potential of our SSR marker and its employment for further research by other in future. While, Sharma and Nandineni, (2014) worked with different Kufri varieties by using same set of SSR markers their study revealed more polymorphism (80- 90%) then the present study. Biniam et al., (2016) found that 97.8% SSR markers were highly polymorphic with an average PIC value of 0.87 which was very promising for the characterization of potato genotypes. In recent times, Duan et al., (2019) used 20 SSR markers for the analysis of genetic diversity among 217 potato cultivars and reported 97.99% polymorphism and 0.83 PIC value which were higher as compare to the current investigation tion. The molecular markers with high PIC values can be utilized for the diversity analysis in potato germplasms.
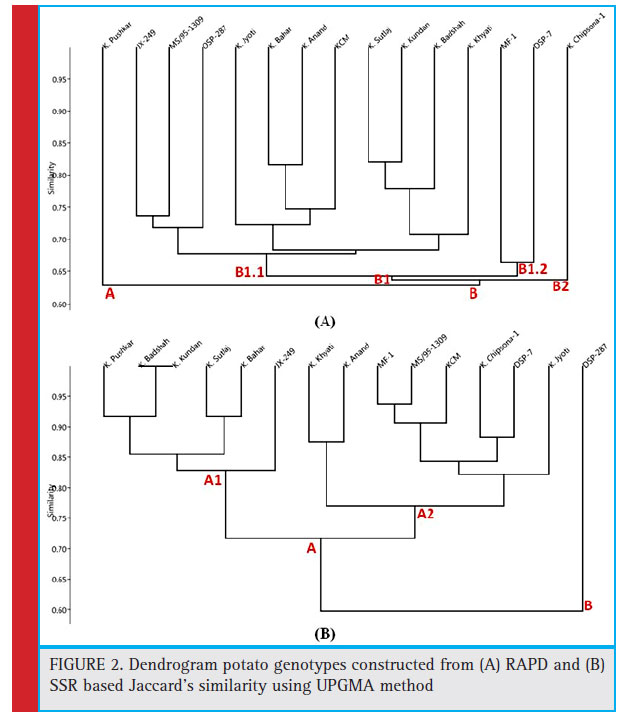 |
Figure 2: Dendrogram potato genotypes constructed from (A) RAPD and (B) SSR based Jaccard’s similarity using UPGMA method |
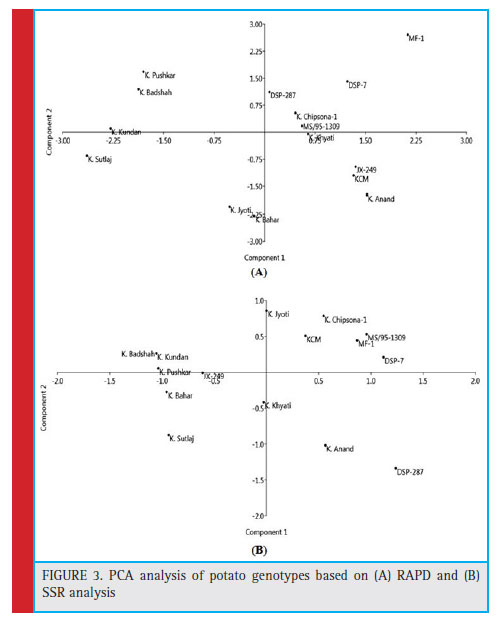 |
Figure 3: PCA analysis of potato genotypes based on (A) RAPD and (B) SSR analysis |
Conclusion
The results of current study indicate that RAPD and SSR markers used in the study seemed to be the good for the molecular assay for fi ngerprinting and assessing genetic relationship among genotypes of potato as they have very promising polymorphism and PIC values. These markers can be utilized for molecular breeding for the improvement of potato.
Acknowledgement
The authors thank authorities of SDAU and staff of Potato Research Station, Deesa, Gujarat, India, for providing facilities to carry out the research work.
References
Ahmed, S., Zhou, X., Pang, Y., Xu, Y., Tong, C., Bao, J. (2018) Genetic diversity of potato genotypes estimated by starch physicochemical properties and microsatellite markers. Food Chemistry 257: 368-375.
Anderson, J.A. (1993) Optimizing parental selection for genetic linkage maps. Genome 36: 181-186.
Ayman, E.F., Zakia, A., Mohamed, T., Sobieh, S., Salah, A. (2018) Molecular diversity analysis of two in vitro and irradiated potato varieties expressed by random amplifi ed polymorphic DNA. Notulae Scientia Biologicae 10(1): 45-51.
Biniam, M.G., Githiri, S.M., Mehari, T., Kasili, R.W., Ghislain, M., Magembe, E. (2016) Genetic diversity assessment of farmers’ and improved potato (Solanum tuberosum) cultivars from Eritrea using simple sequence repeat (SSR) markers. African Journal of Biotechnology 15(35): 1883- 1891.
Bornet, B., Goraguer, F., Joly, G., Branchard, M. (2002) Genetic diversity in European and Argentinian cultivated potatoes (Solanum tuberosum subsp. tuberosum) detected by inter-simple sequence repeats (ISSRs). Genome 45(3): 481-484.
Bradshaw, J.E. (2019) Improving the nutritional value of potatoes by conventional breeding and genetic modifi cation. In Quality Breeding in Field Crops (pp. 41-84). Springer, Cham.
Braun, A., Wenzel, G. (2004) Molecular analysis of genetic variation in potato (Solarium tuberosum L.). I. German cultivars and advanced clones. Potato Research 47: 81-92.
Chimote, V.P., Pattanayak, D., Naik, P.S. (2007) Molecular and morphological divergence studies in Indian potato varieties. Indian Journal of Biotechnology 6: 216-223.
Dharajiya, D.T., Khadia, S.M., Pagi, N.K., Khatrani, T.J., Jasani, H.V., Khunt, A.D., Ravindrababu, Y. (2017) Modifi ed method of high quality genomic DNA extraction from mungbean [Vigna radiata (L.) Wilczek] suitable for PCR based amplifi -cation. Indian Journal of Science and Technology 10(20):1-7.
Doyle, J.J., Doyle, J.L., (1990) Isolation of plant DNA from fresh tissue. Focus 12: 13–15.
Duan, Y., Liu, J., Xu, J., Bian, C., Duan, S., Pang, W., Hu, J., Li, G., Jin, L. (2019) DNA fi ngerprinting and genetic diversity analysis with simple sequence repeat markers of 217 potato cultivars (Solanum tuberosum L.) in China. American Journal of Potato Research 96(1): 21-32.
El Komy, M.H., Saleh, A.A., Molan, Y.Y. (2012) Molecular characterization of early blight disease resistant and susceptible potato cultivars using random amplifi ed polymorphic DNA (RAPD) and simple sequence repeats (SSR) markers. African Journal of Biotechnology 11(1): 37-45.
Favoretto, P., Veasey, E.A., Melo, P.C.T.D. (2011) Molecular characterization of potato cultivars using SSR markers. Horticulture Brasileria 29(4): 542-547.
Feingold, S., Lloyd, J., Norero, N., Bonierbale, M., Lorenzen, J. (2005) Mapping and characterization of new EST-derived microsatellites for potato (Solanum tuberosum L.). Theoretical and Applied Genetics 111(3): 456-466.
Ghebreslassie, B.M., Githiri, S.M., Mehari, T., Kasili, R.W., Ghislain, M., Magembe, E. (2016) Genetic diversity assessment of farmers’ and improved potato (Solanum tuberosum) cultivars from Eritrea using simple sequence repeat (SSR) markers. African Journal of Biotechnology 15(35): 1883-1891.
Ghislain, M., Andrade, D., Rodríguez, F., Hijmans, R.J., Spooner, D.M. (2006) Genetic analysis of the cultivated potato Solanum tuberosum L. Phureja group using RAPDs and nuclear SSRs. Theoretical and Applied Genetics 113(8): 1515-1527.
Ghislain, M., Núnez, J., del Rosario Herrera, M., Pignataro, J., Guzman, F., Bonierbale, M., Spooner, D.M. (2009) Robust and highly informative microsatellite-based genetic identity kit for potato. Molecular Breeding 23(3): 377-388.
Gorji, A.M., Poczai, P., Polgar, Z., Taller, J. (2011) Effi ciency of arbitrarily amplifi ed dominant markers (SCoT, ISSR and RAPD) for diagnostic fi ngerprinting in tetraploid potato. American. Journal of Potato Research 88(3): 226-237.
Hammer, Ø., Harper, D.A.T., Ryan, P.D. (2001) PAST: palaeontological statistics software package for education and data analysis. Palaeontologia Electronica 4(1): 9.
Hawkes, J.G., Jackson, M.T. (1992) Taxonomic and evolutionary implications of the endosperm balance number hypothesis in potatoes. Theoretical and Applied Genetics 84: 180-185.
Jaccard, P. (1908) Nouvelles recherches sur la distribution fl orale. Bulletin de la Société vaudoise des sciences naturelles 44: 223-270.
Jian, W., Lu, H., Wang, R.Y., He, M.M., Liu, Q.C. (2017) Genetic diversity and population structure of 288 potato (Solanum tuberosum L.) germplasms revealed by SSR and AFLP markers. Journal of Integrative Agriculture 16(11): 2434-2443.
Kapuria, M., Dharajiya, D., Khatrani, T., Jasani, H., Chaudhari, S.M., Chauhan, R.M. (2016) Evaluation of Indian potato (Solanum tuberosum L.) germplasms against common scab caused by Streptomyces scabies. International Journal of Agriculture Sciences 8(19): 1336-1338.
Majeed, A., Muhammad, Z. (2018) Potato production in Pakistan: challenges and prospective management strategies–a review. Pakistan Journal of Botany 50(5): 2077-2084.
Maras, M., Sedlar, A., Reid, A., Božovi´c, V., Jovovi´c, Z., Megliˇc, V., Dolniˇcar, P. (2017) Genetic diversity and redundancy among potato accessions in the Montenegrin collection as revealed by microsatellite markers. American Journal of Potato Research 94(4): 306-313.
McGregor, C.E., Lambert, C.A., Greyling, M.M., Louw, J.H., Warnich, L. (2000) A comparative assessment of DNA fi ngerprinting techniques (RAPD, ISSR, AFLP and SSR) in tetraploid potato (Solanum tuberosum L.) germplasm. Euphytica 113(2):135-144.
Oliveira, E.J., Pádua, J.G., Zucchi, M.I., Vencovsky, R., Vieira, M.L.C. (2006) Origin, evolution and genome distribution of microsatellites. Genetics and Molecular Biology 29(2): 294-307.
Onamu, R., Legaria, J., Rodríguez, J.L., Sahagùn, J., Pèrez, J. (2016) Molecular characterization of potato (Solanum tuberosum L.) genotypes using random amplifi ed polymorphic DNA (RAPD) and inter simple sequence repeat (ISSR) markers. African Journal of Biotechnology 15(22): 1015-1025.
Pal, B.P., Nath, P. (1951) Indian potato varieties. Indian Council Agricultural Research, New Delhi. Miscellaneous Bulletin. 62: 63.
Pandey, S.K., Sarkar, D. (2005) Potato in India: Emerging trends and challenges in the new millennium. Potato Journal 32(3-4): 93-104.
Parita, B., Kumar, S.N., Darshan, D., Karen, P. (2018) Elucidation of genetic diversity among ashwagandha [Withania somnifera (L.) Dunal] genotypes using EST-SSR markers. Research Journal of Biotechnology 13(10): 52-59.
Rocha, E.A., Paiva, L.V., Carvalho, H.H.D., Guimarães, C.T. (2010) Molecular characterization and genetic diversity of potato cultivars using SSR and RAPD markers. Crop Breeding and Applied Biotechnology 10(3): 204-210.
Sharma, V., Nandineni, M.R. (2014) Assessment of genetic diversity among Indian potato (Solanum tuberosum L.) collection using microsatellite and retrotransposon based marker systems. Molecular Phylogenetics and Evolution 73: 10-17.
Smith, J.S.C., Chin, E.C.L., Shu, H., Smith, O.S., Wall, S.J., Senior, M.L., Mitchell, S.E., Kresovich, S., Ziegle, J. (1997) An evaluation of the utility of SSR loci as molecular markers in maize (Zea mays L.): comparisons with data from RFLPs and pedigree Theoretical and Applied Genetics 95: 163-173.
Solano Solis, J., Morales Ulloa, D., Anabalón Rodríguez, L. (2007) Molecular description and similarity relationships among native germplasm potatoes (Solanum tuberosum ssp. tuberosum L.) using morphological data and AFLP markers. Electronic Journal of Biotechnology 10(3): 436-443.
Tiwari, J.K., Ali, N., Devi, S., Kumar, V., Zinta, R., Chakrabarti, S.K. (2018) Development of microsatellite markers set for identification of Indian potato varieties. Scientia Horticulturae 231: 22-30.
Vanishree, G., Patil, V.U., Kardile, H., Bhardwaj, V., Singh, R., Chakrabarti, S.K. (2016) DNA fi ngerprinting of Indian potato cultivars by inter simple sequence repeats (ISSRs) markers. Potato Journal 43(1): 70-77.
Walunjkar, B., Parihar, A., Chaurasia, P., Pachchigar, K., Chauhan, R.M. (2013) Genetic analysis of wild and cultivated germplasm of pigeonpea using random amplifi ed polymorphic DNA (RAPD) and simple sequence repeats (SSR) markers. African Journal of Biotechnology 12(40): 823-5832.
Wang, Y., Rashid, M., Li, X., Yao, C., Lu, L., Bai, J., Li, Y., Xu, N., Yang, Q., Zhang, L., Bryan, G.J. (2019) Collection and evaluation of genetic diversity and population structure of potato landraces and varieties in China. Frontiers in Plant Science 10: 139.
Yasmin, S., Islam, M.S., Kondoker, M., Nasiruddin, M., Alam, S. (2006) Molecular characterization of potato germplasm by random amplifi ed polymorphic DNA markers. Biotechnology 5(1): 27-31.


