1Parasitology & Microbiology Research Laboratory, Department of Zoology, The University of Burdwan, Burdwan, West Bengal, India.
Corresponding author email: soumen.microbiology@gmail.com
Article Publishing History
Received: 05/05/2020
Accepted After Revision: 20/06/2020
Starch degrading enzyme amylase has manifold importance with enormous industrial application. Although it can be obtained from various sources, bacteria are the most imperative sources for amylase production. The objective of the present research work is to isolate and characterize bacteria from the gut of earthworm (Perionyx excavatus) those having the ability to degrade starch. In addition, the amylase production by the bacterial isolate EGB3 is determined quantitatively. The bacterial isolate was gram positive, rod shaped, and spore former. EGB3 was found to be non pathogenic and could ferment a wide range of sugars like lactose, xylose, ducitol, sorbitol, galactose, arabinose, inositol, salicin, trehalose, dextrose, fructose, maltose, mannose, sucrose, and cellobiose. The isolate EGB3 produced large halo-zone on starch agar medium and starch degrading index was 2.43. Enzyme production by the isolate EGB3 was 29.45U/mL after 24 h growth in 1% starch broth at 370 C. The bacterial isolate also able to hydrolyze protein (gelatin) and lipid. From the morphological, biochemical and 16S rRNA gene sequence the isolate EGB3 was identified as Bacillus tequilensis.
Starch hydrolysis, Amylase, Amylolytic bacteria, Bacillus tequilensis, Characterization, Earthworm
Ghosh S, Chatterjee S. Isolation and Characterization of Bacillus tequilensis from Gut Content of Perionyx excavatus and Evaluation of its Starch Hydrolyzing Property. Biosc.Biotech.Res.Comm. 2020;13(2).
Ghosh S, Chatterjee S. Isolation and Characterization of Bacillus tequilensis from Gut Content of Perionyx excavatus and Evaluation of its Starch Hydrolyzing Property. Biosc.Biotech.Res.Comm. 2020;13(2). Available from: https://bit.ly/2YPZYzi
Copyright © Ghosh and Chatterjee This is an open access article distributed under the terms of the Creative Commons Attribution License (CC-BY) https://creativecommns.org/licenses/by/4.0/, which permits unrestricted use distribution and reproduction in any medium, provide the original author and source are credited.
INTRODUCTION
Microorganisms especially bacteria have huge importance in production of industrial enzymes (Sadhu and Maiti, 2013; Bharathi et al., 2019). Amylase is one such enzyme which has manifold importance with enormous application in food, paper, fermentation and textile industries (Mishra and Behera, 2008). Amylases constitute a group of industrial enzymes occupying roughly 25% of the marketable enzymes (Sidhu et al., 1997). Although it can be obtained from various sources, bacteria are the most imperative sources for amylase production (Kathiresan and Manivannan, 2006). But not only for the industrial use, the enzyme amylase could be a potential tool in organic waste management. Along with escalating industrialization, urbanization and economic growth, production of solid waste around the Globe is also increasing. Recently, it is assumed to increase the world’s solid waste production up to three billion tons by the year of 2025 (Charles et al., 2009).
According to Pappu et al. (2007) in India, 960 million tons solid wastes are formed each year. Under this circumstance, waste management has become a massive problem and one of the most challenging issues now. Organic wastes from agro-based industries and households chiefly constitute with huge quantities of starch (Jesse et al., 2002). This massive waste can be degraded by the application of amylolytic bacteria. There are diverse reports on starch hydrolyzing microorganisms from various sources (Nimisha et al., 2019).
Earthworms are voracious feeder of organic compounds and their gut content hold a diverse community of microorganisms which are able to degrade macromolecule like starch (Parthi et al., 2019). Indeed, the microorganisms residing in earthworm’s gut are responsible for degradation of organic matter and thus they help in production of vermicompost. Sereval species of Bacillus like Bacillus subtilis, B. Stearothermophilus, B. myocodes , Bacillus amyloliquefaciens, B. polymyxa, B. licheniformis, B. gavealeus, B.mesentericus, , B.vulgates, B. aterrimus were reported as amylase producer.
Especially, Bacillus subtilis, B.licheniformis and Bacillus amyloliquefaciens have been reported to produce adequate quantity of alpha amylase (Singh et al., 2011). During present study, starch hydrolyzing Bacillus tequilensis (EGB3) isolated from earthworm gut has been characterized by morphological, biochemical and molecular methods and identified through polyphasic methods. The starch hydrolyzing ability of the bacterial isolate was determined both qualitatively as well as quantitatively.
MATERIAL AND METHODS
Live earthworms (Perionyx excavatus) used in vermicompost preparation, were collected in sterile plastic container from Kulti vermicomposting farm (23°12′ N, 88°30′ E) of Purba Bardhaman district of West Bengal, India. After collection, earthworms were brought to Parasitology and Microbiology Research Laboratory of the Department of Zoology, the University of Burdwan for further microbial and biochemical analysis.
Live earthworms were surface sterilized with 50% alcohol and quickly transferred to a dissection tray. The gut was cut open and the gut content was collected with a sterile loop into an Eppendorf tube. The semi solid gut content was diluted with sterile distilled water and pour plated on Nutrient Agar (NA) medium (Himedia, India) [Composition: peptone 5.0, Yeast extract 1.5, HM peptone 1.5, NaCl 5.0, agar 15 g l-1]. The plates were incubated at 30±10C for 24-48h. At the end of incubation, bacterial colonies were picked up from plates and streaked repeatedly on NA plate and maintained in pure culture at 40C for further study.
The morphological and biochemical characterization of bacterial isolate were done following standard methods (Holt, 1984; Smibert et al. 1995; Logan et al., 2009). The Gram staining was performed using Gram staining kit (Himedia, India). SIM agar was used to perform the motility test of the bacterial isolate by stabbing into the slant (Czaban et al., 2007). The biochemical characteristics such as Methyl-Red (MR), catalase, Voges-Proskauer (VP), citrate utilization, nitrate reduction, indole production, urease, oxidase, and carbohydrate fermentation tests were performed. For the determination of pathogenic nature the isolate blood hemolysis and DNase test were conducted (Benson et al., 2012).
To prepare a scanning electron micrograph a thin smear was made on cover slip, fixed using 2.5% glutaraldehyde and gradually dehydrated with graded alcohol. Finally the specimen was gold coated and observed under scanning electron microscope (ZEISS). Antibiotic sensitivity test was performed with standard antibiotics discs (Himedia, India) following Brown (2004) and sugar fermentation test was carried out with sugar fermentation discs like inositol, maltose, sucrose, adonitol, fructose, mannose, cellobiose, raffinose, xylose, dextrose, galactose, lactose, trehalose, rhamnose, ducitol, melibiose, , salicin, sorbitol, mannitol, and arabinose (Himedia, India).
The sensitivity of the isolate against antibiotics was tested using nalidixic acid, kanamycin, tetracycline, ofloxacin, ampicillin, amoxicillin, neomycin, gentamycin, ciprofloxacin, azithromycin, erythromycin, bacitracin, chloramphenicol, penicillin, rifampicin, and doxycycline. The zone diameter of inhibition (ZDI) values were interpreted following CLSI (2011). Multiple Antibiotic Resistance (MAR) index were calculated following Krumperman (1983).
Number of antibiotics to which the isolate showed resistance
MAR index = ——————————————————————————
Number of total antibiotics exposed to the isolate
For molecular characterization, Genomic DNA of bacterial isolate was extracted using DNeasy Ultra Clean Microbial Kit of Qiagen. Fragment of ~1.5 kb rDNA was amplified by Polymerase Chain Reaction (First cycle at 940C, thirty five cycles at 580C and finally at 720C) using 27F forward (5′AGAGTTTGATCATGGCTCAG 3′) and 1492 reverse (5′GGT TAC CTT GTT ACG ACTT3′) primer. Then the PCR product was purified with MinElutePCR purification kit, Qiagen. Purified PCR product was sequenced using universal bacterial primer in DNA sequencer. Sequenced information was aligned by ClustalW (http://www.ebi.ac.uk/clustalw) and analyzed using MEGA 4.0.2 software (Thompson et al., 1994). Evolutionary distances of mostly related bacteria were calculated following the method of Jukes and Cantor 1969 and phylogenetic tree of the bacterial isolate was constructed following Tamura et al., (2007) 1.
For screening of amylolytic bacteria starch hydrolysis test was performed using iodine and starch agar (Himedia, India) [Meat Extract 3.0; Peptic digest of animal tissue 5.0; Starch, soluble 2.0; Agar 15.0 g l-1; pH 7.2±0.1]. Colonies of the bacterial isolate were transferred on to starch agar plates and incubated at 37±1oC for 48 h. Then the plate with bacterial colony was flooded with Gram’s iodine. If a isolate was amylolytic then it would produce a clear zone around the colony. This clear zone is formed as enzyme amylase starts hydrolyzing the starch molecule. Selection was made as per colonies with and without clear and transparent zone as starch hydrolyzing and starch non-hydrolyzing isolate, respectively.
The Hydrolyzing Capacity of the bacterial isolate was determined by Starch Degrading Index (SDI) and it was indexed as the diameter of the colony plus the clear zone around it divided by the diameter of the colony (Nusrat and Rahman, 2007). The largest ratio represented the highest activity. The amylase assay was carried out using 3-5-dinitrosalicylic acid reagent (DNS Reagent) (Bernfeld, 1955).
The bacterial isolate was inoculated in 20ml Minimal Starch Broth and incubated in the shaker incubator for 24 hours at 30oC at 200rpm. 1ml of this broth was then inoculated in 100ml of minimal broth containing 1% starch and incubated in shaker incubator at 200rpm for 48 hours at 30 ºC. There after the culture was centrifuged at 2500 rpm. The supernatant was used as a source of crude enzyme extract. 1 ml of bacterial crude enzyme was taken in a test tube containing 2 ml of 1% starch broth. Then the reaction mixture was incubated at 50ºC for 60 minutes. To arrest the enzyme action, 1ml of DNS reagent was added. The test tubes were kept in boiling condition upon water bath for 5 minutes. 1 ml of 40% sodium potassium tartarate was then added into the test tubes and allowed to cool. The optical density of the tubes was measured at 540 nm. Enzyme activity was expressed in units (Mishra and Behera, 2008).
RESULTS AND DISCUSSION
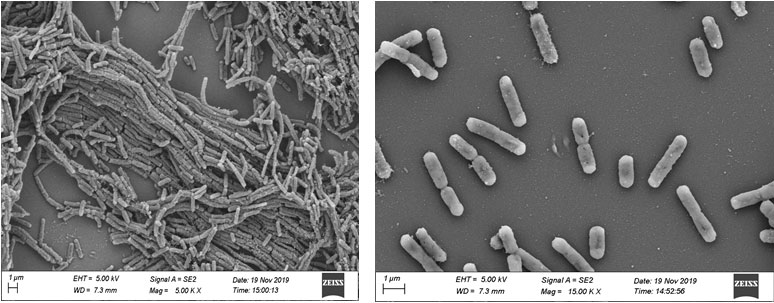
The colony of the bacterial isolate EGB3 appeared on NA plate was rounded in shape with a diameter of 3-3.5 mm, moderately elevated with smooth margin, opaque and offwhite in colour. EGB3 was a Gram positive, spore former, rod shaped bacterium with a length of 784.6-802.5 nm and breadth of 3.378-3.423 µm (Figure 1). The motility test of the isolate indicated its motile character. EGB3 was found to be non pathogenic against blood hemolysis test and DNase test. The bacterial isolate was able to produce enzyme catalase, oxidase and utilized citrate as carbon source, but it was unable to produce urease. The isolate was positive for both MR and VP test. EGB3 was able to degrade arginine, tryptophan and positive for ONPG test. The isolate EGB3 could hydrolyze gelatin, lipid as well as starch (Table 1).
Table 1. Morphological and biochemical characteristics of isolated strain Bacillus tequilensis EGB3.
| Test | Response |
| Gram’s staining | Gram positive |
| Shape | Rod |
| Length of bacterium | 784.6-802.5 nm |
| Breadth of bacterium | 3.378-3.423 µm |
| Motility | Positive |
| MR | Positive |
| VP | Positive |
| Catalase | Positive |
| Citrate | Positive |
| Urease | Negative |
| Oxidase | Positive |
| H2S Production | Negative |
| Blood hemolysis | Negative |
| DNase test | Negative |
| Indole | Positive |
| Nitrate Reduction to N2 | Positive |
| Starch hydrolysis | Positive |
| Protein (gelatin) hydrolysis | Positive |
| Fat hydrolysis | Positive |
Figure 1: Scanning microscopy of Bacillus tequilensis (EGB3) showing single cell and chain formation by multiple bacteria.
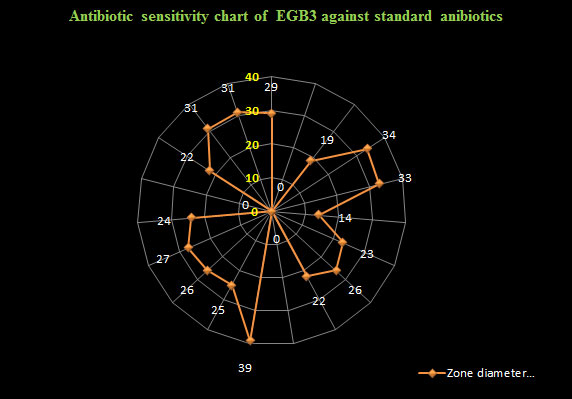
The bacterial isolate (EGB3) showed the ability to ferment inositil, galactose, xylose, salicin, trehalose, rhamnose, arabinose, dextrose, dulcitol, sorbiol, fructose, maltose, mannose, sucrose, lactose and cellobiose (Table 2). Bacterial isolate EGB3 showed sensitivity to the recommended doses of antibiotics like tetracycline (30 µg/ disc), ofloxacin (5 µg/ disc), doxycycline (30 µg/ disc), nalidixic acid (30 µg/ disc), erythromycin (15 µg/disc), neomycin (30 µg/disc), amoxicillin (10 µg/ disc), bacitracin (10/disc µg), levofloxacin (5 µg/ disc), streptomycin (10 µg/ disc), azithromycin (30 µg/ disc), gentamycin (50 µg/ disc), vancomycin (30 µg/ disc), chloramphenicol (30 µg/ disc), kanamycin (30 µg/ disc) and norfloxacin (10 µg/disc) but resistance to rifampicin (5 µg/ disc), ampicillin (10 µg/ disc) and penicillin (10/disc µg) (Figure 2) . MAR index of EGB3 was 0.15.
Table 2. Carbohydrate fermentation property of Bacillus tequilensis EGB3.
| Carbohydrate | Response |
| Inositol | Positive |
| Salicin | Positive |
| Trehalose | Positive |
| Galactose | Positive |
| Xylose | Positive |
| Dextrose | Positive |
| Adonitol | Negative |
| Lactose | Positive |
| Arabinose | Positive |
| Melibiose | Negative |
| Fructose | Positive |
| Maltose | Positive |
| Mannose | Positive |
| Sucrose | Positive |
| Rhamnose | Positive |
| Cellobiose | Positive |
| Mannitol | Negative |
| Ducitol | Positive |
| Raffinose | Negative |
| Sorbitol | Positive |
Figure 2: Antibiotic sensitivity and ZDI values of the isolate EGB3 against standard antibiotics.
The bacterial isolate (EGB3) was able to hydrolyze starch as it produced large clear zone around the colony (Figure 3). The SDI calculated from the clear zone and colony ratio was found to be 2.43 during the current study. Enzyme production by the isolate EGB3 was 29.45U/mL after 24 h cultivation in 1% starch broth at 370C.
Figure 3: The amylase production by Bacillus tequilensis EGB3.
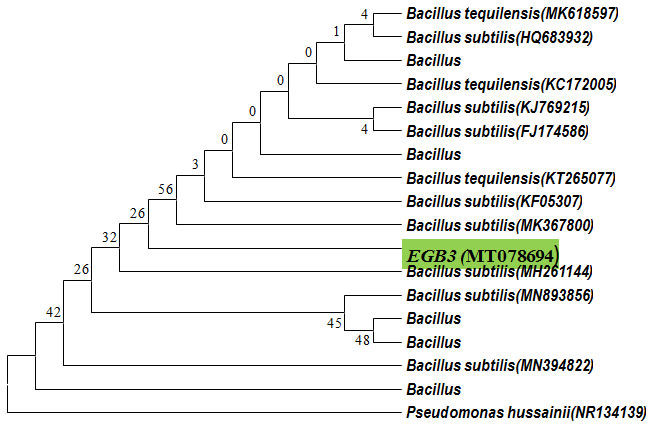
Neighbour joining tree has been constructed by partial 16S rRNA sequence of the isolate using Pseudomonas hussainii as an out-group (Figure 4). From Neighbour joining tree it was found that the bacterium EGB3 clustered with both Bacillus subtilis and Bacillus tequilensis [Bacillus tequilensis (MK618597; HQ238557; KC172005; HQ238430; KT265077) Bacillus subtilis (HQ683932; KJ769215; FJ174586; KF05307; MK367800]. Gatson et al. (2006) previously reported that there was 99% similarity in 16S rRNA gene sequences of B. tequilensis and B. subtilis. From the16S rDNA gene sequencing it was found that EGB3 might be a strain of B. tequilensis or B. subtilis. But the biochemical properties like enzymatic and fermentation properties of EGB3 shows higher affinities with B. tequilensis. For example, EGB3 and B. tequilensis both could ferment galactose, rhamnose, lactose and dulcitol but B. subtilis could not ferment these sugars. In addition, unlike to B. subtilis, EGB3 was able to decompose arginine and tryptophan. B. tequilensis was also able to degrade arginine, tryptophan and ONPG (Gatson et al., 2006).
Figure 4: Phylogenetic neighbor joining tree constructed based on partial 16S rRNA gene sequence of EGB3 (MT078694) strain along with the other 16S rRNA gene sequnces retrieved from NCBI and RDP.
Moreover, other biochemical properties like MR, VP, indole and catalase production, citrate and urea utilization, starch and gelatine hydrolysis strongly suggested the similarity between isolate EGB3 and previously reported B. tequilensis (Li et al., 2018; Gatson et al., 2006) . The percentage of AT and GC content of 16S rRNA gene sequence of EGB3 were 44% and 56% respectively. From the morphological, biochemical and molecular analysis the isolate EGB3 was identified as B. tequilensis.
Vermicompost harbours a rich variety of microbial populations such as bacteria, fungi and actinobacteria (Edwards, 1998; Samanta and Das, 2016). Role of earthworms and microbes in decomposition and humification of organic substances has been studied by various researchers (Edwaeds and Bohlen. 1996; Cai et al., 2002; Manivannan et al., 2004; Munnoli and Bhosle, 2014). In the present study, it was found that the gut of Perionyx excavatus contained bacteria that could produce amylase, a starch hydrolysing enzyme. Enzyme α-amylase belongs to a group of endo-amylases which catalysed the hydrolysis of starch into shorter oligosaccharides through the cleavage of α-D-(1-4) glycosidic bonds (Souza et al., 2010). Although it could be obtained from various sources, bacteria were the most imperative sources for amylase production (Gangadharan et al., 2006). The microbial amylase has a wide range of industrial applications (Mishra and Behera, 2008).
Amylases of microbial origins are more stable than amylase of animal or plant origin (Tanyildiz iand Elibol, 2005). Moreover, the foremost advantage of using microorganisms for amylase production is the cost-effective mass production capacity and as microbes are uncomplicated to manipulate, enzymes of desired characteristics are obtained easily (Souza et al., 2010). Industrially produced first enzyme was an amylase from fungi in 1894, which was used in making of a pharmaceutical aid (Crueger and Crueger, 1989; Pandey et al., 2000). Biodin and Effront were the pioneers for the production of α-amylase on commercial scale from B. subtilis and B. mesentericus. (Hoogerheide, 1954).
In this present study the amylolytic bacterial isolate was identified and this result is similar to the findings by many workers (Parthasarathi et al., 2007; Samanta and Das, 2016). Several species of Bacillus like Bacillus subtilis, B. stearothermophilus, B. myocodes , Bacillus amyloliquefaciens, B. polymyxa, B. licheniformis, B. gavealeus, B.mesentericus, , B.vulgates, B. aterrimus were reported as amylase producer. Especially, Bacillus subtilis, B.licheniformis and Bacillus amyloliquefaciens have manifold importance in commercial applications as they produce adequate quantity of alpha amylase (Singh et al., 2011). B. tequilensis was foremost reported by Gatson et al in 2006 from two thousand year-old Mexican shaft-tomb near the city of Tequila. Kaur and Azmi (2013) found B. tequilensis from earthworm’s intestinal content which have high collagenase activity. B. tequilensis was previously reported as starch hydrolyzer ( Prasanth et al., 2017 Li et al 2028).
Mishra and Behera (2008) reported amylolytic Bacillus spp. from kitchen waste. The kitchen wastes, predominantly consists of starchy materials and they advocated that bacteria isolated from such places possessed better potential to produce amylase under adverse situation. Amylase production efficiency by the bacteria of same genus even of the same species differs significantly. A branch of factors regulates the production of amylase. Temperature, pH, growth kinetics of isolates, incubation time, size of inoculums, Carbon sources, etc. play important roles in production of bacterial amylase, (Alariya et al., 2013). In an experiment on starch hydrolysis by amylase was exhibited by Dida (2018), who observed that the SDI of rhizospheric Bacillus spp. ranged between 1.23 and 2.15. The starch hydrolyzing ability from present research work in relation to SDI was comparable with Bacillus spp. (Chatterjee et al., 2019).
According to Pappu et al., (2007) in India, 960 million tons solid waste is produced each year. Under this circumstance, waste management has become a massive problem and one of the most important issues now. Present study showed that the gut bacteria of Perionyx excavatus may be a potential tool in removal of agricultural waste through its starch hydrolysing activity. This study is at preliminary level and it requires some more research for better understanding of diversity and potential application of the earthworm gut bacteria in sustainable agricultural practice, organic waste management and other land uses.
CONCLUSION
From the present study it is found that the gut content of earthworm contains amylolytic bacteria like Bacillus tequilensis which have a great potentiality to degrade starch. The earthworm Perionyx excavatus, or the bacteria itself may be utilized in organic waste management and the amylolytic property of the isolate can be explored commercially in various aspects.
ACKNOWLEDGEMENTS
The authors are grateful to UGC, DST-FIST and DST-PURSE for providing instrumental facilities. The authors are thankful to the Head, Department of Zoology, and the University of Burdwan for giving all sorts of laboratory facilities to conduct this research. We are also grateful to Hon’ble Vice Chancellor, the University of Burdwan, for his moral support and inspiration.
Conflict of Interest: The authors declare that they have no conflict of interest.
REFERENCES
Alariya SS, Sethi S, Gupta S, Lal GB, Lal GB (2013) Amylase activity of a starch degrading bacteria isolated from soil. Arch Appl Sci 5(1):15-24.
Benson DA, Karsch-Mizrachi I, Clark K, Lipman DJ, Ostell J, Sayers EW (2012) GenBank. Nucleic Acids Res 40:48-53.
Bernfeld P (1955) α-and β-amylases. Methods Enzymol 1(l.c):149-158.
Bharathi D, Rajalakshmi G, Komathi S (2019) Optimization and production of lipase enzyme from bacterial isolates isolated from petrol spilled soil. J King Saud Univ Sci 31(4): 898-901.
Brown A (2004) Benson’s Microbiological applications: Laboratory Manual in General Microbiology. McGraw-Hill Science/Engineering/Math, New York.
Cai H, Zarda B, Mattison GR, Schönholzer F, Hahn D (2002) Fate of Protozoa transiting the digestive tract of the earthworm Lumbricus terrestris L. Pedobiologia 46(2):161-175.
Charles W, Walker L, Cord-Ruwisch R (2009) Effect of pre-aeration and inoculum on the start-up of batch thermophilic anaerobic digestion of municipal solid waste. Bioresour Technol 100(8):2329-2335.
Chatterjee P, Roy P, Mandal P, Chatterjee S (2019) Isolation, phenotypic and molecular characterization of Burkholderia sp. (isolate, PCS1) from maize fields exhibiting starch hydrolysis ability. Biosci Biotech Res Comm 12(1):66-72.
CLSI (2011) Clincal Laboratory Standards Institute. Performance Standards for Antimicrobial Susceptibility Testing; Twenty-First Informational Supplement. Wayne, Pennsylvania, USA.
Crueger W, Crueger A (1989) Ed. Industrial Microbiology, Sinauer Associates, Sunderland, MA, 189–218.
Czaban J, Gajda A, Wróblewska B (2007). The motility of bacteria from rhizosphere and different zones of winter wheat roots. Pol J Environ Stud 16(2):301-308.
Edward CA (1998) The use of earthworms in the break down and management of organic wastes. In: Edwards CA (ed), Earthworm Ecology. CRC Press LLC, Boca Raton, FL, USA, pp 327–354.
Edwards CA, Bohlen PJ (1996) Biology and Ecology of Earthworms. 3rd edn, Chapman and Hall, London.
Gangadharan D, Sivaramakrishnan S, Nampoothiri KM, Pandey A (2006) Solid culturing of Bacillus amyloliquefaciens for α-amylase production. Food Technol Biotech 44(2):269-274.
Gatson JW, Benz BF, Chandrasekaran C, Satomi M, Venkateswaran K, Hart ME (2006) Bacillus tequilensis sp. nov., isolated from a 2000-year-old Mexican shaft-tomb, is closely related to Bacillus subtilis. Int J Syst Evol Microbiol 56(7):1475-1484.
Holt JG, Krieg NR, Sneath PHA, Staley JT Williams ST (1994) Bergey’s Manual of Determinative Bacteriology (9th Int ed), Williams and Wilkins, Maryland, USA, pp 565- 596.
Hoogerheide JC (1954) Microbial enzymes other than fungal amylases. Industrial Fermentations, Chemical Publishing Co., New York.
Jesse TW, Ezeji TC, Qureshi N, Blaschek HP (2002) Production of butanol from starch-based waste packing peanuts and agricultural waste. J Ind Microbiol Biotechnol 29(3):117-123.
Jukes TH, Cantor CR (1969) Evolution of protein molecules. In: Munro HN (ed). Mammalian Protein Metabolism, New York. Academic Press.
Kathiresan K, Manivannan S (2006) -Amylase production by Penicillium fellutanum isolated from mangrove rhizosphere soil Afr J Biotechnol 5(10):829–832.
Krumperman PH (1983) Multiple antibiotic resistance indexing of Escherichia coli to identify high-risk sources of fecal contamination of foods. Appl Environ Microbiol 46(1):165-170.
Li H, Guan Y, Dong Y, Zhao L, Rong S, Chen W, Li L (2018) Isolation and evaluation of endophytic Bacillus tequilensis GYLH001 with potential application for biological control of Magnaporthe oryzae. PloS one 13(10).
Logan NA, De Vos P (2009). Genus I. Bacillus. Bergey’s manual of systematic bacteriology 3 pp21-128.
Manivannan S, Ramamoorthy P, Parthasarathi K, Ranganathan LS (2004) Effect of sugar industrial wastes on the growth and reproduction of earthworms. India J Exp Zool 7(1):29–37.
Manohar P, Shanthini T, Gothandam KM, Kannan VR, Ramesh N (2017) Enhanced amylolytic activity of intracellular α-amylase produced by Bacillus tequilensis. J Microbiol Biotechnol Food Sci 6(6).
Mishra S, Behera N (2008) Amylase activity of a starch degrading bacteria isolated from soil receiving kitchen wastes. Afr J Biotechnol 7(18):3326-3331.
Munnoli PM, Bhosle S (2014) Role of bacterial Inoculum. Proceedings of International Conference on Solid Waste Technology organized by Wiener University, Philadelphia, USA, March 2014, pp1339–1360.
Nimisha P, Moksha S, Gangawane AK (2019) Amylase Activity of Starch Degrading Bacteria Isolated from Soil. Int J Curr Microbiol App Sci 8(4):659-671.
Nusrat A, Rahman SR (2007). Comparative studies on the production of extracellular a-amylase by three mesophilic Bacillus isolates. Bangladesh J of Microbiol 24(2):129-132.
Pandey A, Soccol CR, Mitchell D (2000) New developments in solid state fermentation. Process Biochem 35:1153-1169.
Pappu A, Saxena M, Asolekar SR (2007). Solid wastes generation in India and their recycling potential in building materials. Build Environ 42(6):2311-2320.
Parhi DK, Sridhar T, Deepthi MP, Kathireswari P (2019) Isolation And Characterization Of Gut Bacterial Species Of Earthworm Lampito mauritii. Kongunadu Res J 6(2):45-50.
Parthasarathi K, Ranganathan LS, Anandi V, Zeyer J (2007) Diversity of microflora in the gut and casts of tropical composting earthworms reared on different substrates. J Environ Biol 28(1):87–97.
Sadhu S, Maiti TK (2013) Cellulase production by bacteria: a review. Microbiol Res J Int 235-258.
Samanta TT, Das A (2016) Isolation, identification, and characterization of gut microflora of Perionyx excavatus collected from Midnapore, West Bengal. J Basic Microbiol 56(3):286-293.
Sidhu GS, Sharma P, Chakrabarti T, Gupta, JK (1997) Isolate improvement for the production of a thermostable α-amylase. Enzyme Microb Technol 21(7):525-530.
Singh S, Sharma V, Soni ML, Das S (2011) Biotechnological applications of industrially important amylase enzyme. Int J Pharma Bio Sci 2(1):486-496.
Smibert R, Krieg NR (1995). Phenotypic testing. In Methods for General and Molecular Bacteriology. American Society for Microbiology, pp 607-654.
Smits JP, Rinzema J, Tramper H, Van M, Knol W (1996) Solid-state fermentation of wheat bran by Trichoderma reesei QM9414: substrate composition changes, C balance, enzyme production, growth and kinetics. Appl Micro Biotechnol 46:489-496.
Sneath PHA, Holt JG (2001) Bergey’s Manual of Systematic Bacteriology (2nd ed.) Williams and Wilkins, Springer-Verlag, NY, USA, 1, pp 64.
Souza PMD (2010) Application of microbial α-amylase in industry-A review. Brazilian J. Microbial., 41(4):850-861.
Tanyildizi MS, Özer D, Elibol M (2005) Optimization of α-amylase production by Bacillus sp. using response surface methodology. Process Biochem 40(7):2291-2296.
Thompson JD, Higgins DG, Gibson TJ (1994). Clustal W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, positionspecifi c gap penalties and weight matrix choice. Nucleic Acids Res, 22: 4673-4680.