Department of Zoology, University of Gour Banga, Malda-732103, India
Corresponding author Email: samtropmed@gmail.com
Article Publishing History
Received: 01/06/2018
Accepted After Revision: 10/08/2018
This communication characterizes the curd isolate of lactic acid bacteria having the capacity to antagonize human pathogenic bacteria. The commercially available curd, in sealed form in a plastic cup, was procured from Malda town market (West Bengal state, India) and processed microbiologically, using de Man Rogosa Sharpe medium, for the isolation of lactic acid bacteria. The pure bacteria culture obtained was identified, by phenotypic characterization through conventional methods, as Lactobacillus fermentum, and designated as LMEM 22. The Lactobacillus fermentum LMEM 22 curd isolate had mixed antibiotic susceptibility patterns, showing resistance (ZDI: ≤15 mm) to amikacin, ciprofloxacin, kanamycin, methicillin and vancomycin, sensitivity (ZDI: ≥ 21 mm) to ampicillin, amoxyclav, gentamycin, cefotaxime, imipenem, meropenem and tetracycline, and intermediate susceptibility (ZDI: 16 – 20 mm) to cfoxitin and trimethoprim. The L. fermentum LMEM22 antagonizes both gram-negative: Acinetobacter baumannii, Pseudomonas aeruginosa, Escherichia coli, Proteus vulgaris, Klebsiella pneumoniae, Salmonella enterica serovar Typhi, and gram-positive: Staphylococcus aureus, Bacillus cereus, Enterococcus faecalis, Listeria monocytogenes, bacterial pathogens, following agar overlay (ZDI: 17±1.73 mm to 20±1.00 mm, for gram-positive, and 18±2.00 mm to 33±2.65 mm for gram-negative bacteria) as well as agar-well diffusion (ZDI: 10.67±2.08 mm to 12±1.00 mm, for gram-positive, and 13.00±2.65 mm to 18.00±3.00 mm, for gram-negative bacteria) techniques. The overall bacteriocin activity (AU/ml) of Lactobacillus fermentum LMEM 22 for the test bacterial pathogens ranged 142.27 – 240.00, and the ‘R’ value ranged 5.5 – 13.5. This study underlines the usefulness of locally available lactic acid bacteria in designing the probiotic microorganisms for biotherapy.
Lactic Acid Bacteria, Lactobacillus Fermentum, Antagonistic Activity, Bacteriocin Activity, Pathogenic Bacteria
Halder D, Mandal S. Insights into the Antagonism of Lactobacillus Fermentum Curd Isolate Against Gram-Positive and Gram-Negative Pathogenic Bacteria. Biosc.Biotech.Res.Comm. 2018;11(3).
Halder D, Mandal S. Insights into the Antagonism of Lactobacillus Fermentum Curd Isolate Against Gram-Positive and Gram-Negative Pathogenic Bacteria. Biosc.Biotech.Res.Comm. 2018;11(3). Available from: https://bit.ly/2MC4OfD
Introduction
Among the lactic acid bacteria (LAB), Lactobacillus spp. are characteristically known as probiotics, meaning, as per the definition of FAO/WHO (2001), the ‘live microorganisms which when administered in adequate amounts confer a health benefit on the host’; LAB isolates are beneficial in many ways, but, essentially by restricting the toxigenic bacterial growth in the gut (Podolsky, 1998). The FAO/WHO (2007) suggested that the probiotic microorganisms must possess the capacity to display the antagonistic activity against bacterial pathogens. Among the large number of lactobacilli isolated from various fermented foods, 42 isolates showed activity against Escherichia coli, while 15 isolates had antibacterial activity against Klebsiella pneumoniae (Shehata et al., 2016).
Nivien et al. (2016) isolated LAB from fermented milk, identified the isolates by phenotypic characterization and reported their antibacterial activity against the bacterial strains: Escherichia coli, Salmonella Typhimurium, Staphylococcus aureus and Listeria monocytogenes. It has been reported that the lactobacilli, including Lactobacillus fermentum, had growth inhibitory action against gram-positive as well as gram-negative human pathogenic bacteria (Vuotto et al., 2016). As per the report of Sharma et al. (2016), the LAB (Pediococcus acidilactici and Lactobacillus casei) isolated from milk cream and lassi had growth inhibitory activity against a number of gram-positive food borne bacteria.
Benavides et al. (2016) demonstrated that the Lactobacillus fermentum isolate from local ecological niche was sensitive to ampicillin, cefuroxime, tetracycline and amoxicillin/clavulanic acid and resistant to gentamycin and kanamycin, and the LAB was found inhibitory to Escherichia coli and Salmonella Typhimurium. As per the previous report (Halder et al., 2017), four lactobacilli (Lactobacillus animalis LMEM6, Lactobacillus plantarum LMEM7, Lactobacillus acidophilus LMEM8 and Lactobacillus rhamnosus LMEM9) procured from different commercially available curd samples had antibacterial activity against gram-negative pathogenic bacteria, such as Escherichia coli, Proteus vulgaris, Acinetobacter baumannii and Salmonella enterica serovar Typhi, and had resistance to vancomycin and amoxyclav.
Recently, Mahalot and Mandal (2018) have isolated LAB from locally available cow milk and goat milk samples showing sensitivity to most of the test antibiotics, while resistance was recorded for all isolates to methicillin, for Lactobacillus sp. G1 and Lactococcus sp. G2 to trimethoprim, while to vancomycin for Lactobacillus sp. G1 and Lactobacillus sp. C1. Since the good LAB are not even waived from antibiotic resistance phenomenon, many authors documented the status of various resistances to antibiotics for safety profiling of native LAB isolates (Mandal et al., 2017). This background prompted us to assess the broad spectrum antibacterial activity of lactic acid bacillus isolated from commercially available curd, and explore the antibiogram of the isolated Lactobacillus, through phenotypic characterization.
Material and Methods
A single cup of commercially available curd sample was procured from Malda town market (West Bengal, India), and processed microbiologically for the isolation of lactic acid bacteria (LAB), following the protocol mentioned earlier (Halder and Mandal, 2015): growth enrichment of LAB in MRS broth (Hi-Media, India), pure culture (single discrete colony isolation) of LAB on MRS agar (Hi-Media, India) plate, and storage of the LAB in MRS agar stab at 4oC for further processing. The isolated LAB (n=1), following Bergey’s manual (Holt, 1984), as described earlier (Halder and Mandal, 2015), was subjected to phenotypic (gram-staining, colony morphology study and motility test) and biochemical (oxidase and catalase production) characterization. The non-motile non-spore forming gram-positive rod shaped bacteria (no cocci were found), showing negative results to oxidase and catalase tests, were subjected to IMViC, amino acid decarboxylation and sugar fermentation tests.
The antibiotic susceptibility of the LAB (LMEM 22) was executed by disc diffusion method (Bauer et al., 1996), the details of which was described before (Halder and Mandal, 2016; Halder et al., 2017). The antibiotic discs (Hi-Media, Mumbai, India) used in the study included amikacin (Ak: 30-µg/disc), amoxyclav (Ac: 30-µg/disc), ampicillin (Am: 10-µg/disc), ciprofloxacin (Cp: 5-µg/disc), cfoxitin (Cx: 30-µg/disc), cefotaxime (Ct: 30-µg/disc), cefotaxime/clavunilic acid (Cc: 30/10-µg/disc), gentamycin (Gm: 30-µg/disc), imipenem (Ip: 10-µg/disc), kanamycin (Km: 30-µg/disc), methicillin (Me: 5-µg/disc), meropenem (Mp: 10-µg/disc), tetracycline (Tc: 30-µg/disc), trimethoprim (Tm: 5-µg/disc) and vancomycin (Vm: 30-µg/disc). The results, in terms of ZDI (zone diameter of inhibition) values, were interpreted according to Liasi et al. (2009) and Vlkova et al. (2006), in order to label the test bacterial isolate as resistant (ZDI: ≤ 15 mm), sensitive (ZDI: ≥21 mm), or intermediately susceptible (ZDI: 16–20 mm).
The antagonistic activity of the LAB LMEM 22 isolate from curd was determined against gram-negative (Acinetobacter baumannii, Pseudomonas aeruginosa, Escherichia coli, Proteus vulgaris, Klebsiella pneumoniae, and Salmonella enterica serovar Typhi) and gram-positive (Staphylococcus aureus, Bacillus cereus, Enterococcus faecalis, Listeria monocytogenes) bacterial pathogens, following agar-well diffusion (Tagg, 1971; Halder et al., 2017) and agar overlay (Shokryazdan et al., 2018) methods, as described and interpreted earlier (Shokryazdan et al., 2014, Halder et al., 2017, Mandal and Halder, 2018).
The ‘R’ values, from the action of LMEM 22 isolate over the bacterial pathogens, were calculated applying the formula described elsewhere (Halder and Mandal, 2016), and interpreted according to the criteria mentioned earlier (Carasi et al., 2014; Pisano et al., 2014), while the bacteriocin activity of LAB (LMEM 22), in terms of arbitrary units per milliliter (AU/ml), was calculated following the formula put forwarded by Iyapparaj et al. (2013).
Results and Discussion
As has been demonstrated by Iyapparaj et al. (2013), the morphologically identical bacterial colonies, procured from goat milk on the MRS agar plate, have been identified as Lactobacillus sp., on the basis of physical and biochemical characteristics, following Holt et al. (1984). In this study, a single isolate of non-motile non-spore forming gram-positive rod was procured from the curd sample (Figure 1), and the isolated bacteria (LMEM 22) was an hetero-fermentative strain, which in TSI test showed the production of acid as well as gas (CO2). The LMEM 22 isolate showed negative test results for catalase and oxidase, and in IMViC test battery the isolates was positive for methyl red. The sugar fermentation pattern of LMEM 22 isolate is represented in Table 1, while the amino acid decarboxylation test results are depicted in Figure 2. Thus, following phenotypic and biochemical characterization the isolated LAB was identified as Lactobacillus fermentum LMEM 22. The LAB isolates procured from different fermented foods, including curd, have been identified earlier by Nigam et al. (2012), following phenotypic characterization of the bacteria. Currently, the treatment options with antibiotics are inadequate because of the escalating rate of emergence of antibiotic resistant pathogenic bacteria causing life-threatening infections to humans. Alternative to the antibiotics, which remains the mainstay of all therapy for bacterial infections (Van Boeckel et al., 2014), probiotic lactobacilli have been found suitable for biotherapy with proven antibacterial activity (Iyapparaj et al., 2013; Sing et al., 2017).
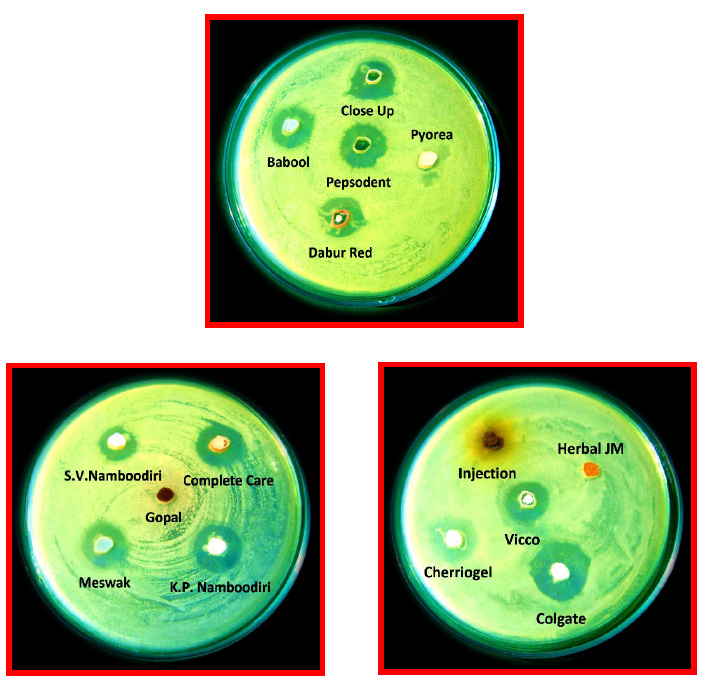 |
Figure 2: Amino acid decarboxylation test results for the isolated LAB from curd sample. The LAB utilized arginine, but not ornithine and L-lysine, in Moeller decarboxylase broth (Hi-Media, India). |
Earlier, it has been reported that the curd isolates of Lactobacillus animalis LMEM6, Lactobacillus plantarum LMEM7, Lactobacillus acidophilus LMEM8 and Lactobacillus rhamnosus LMEM9, had bacterial growth inhibitory activity, having ZDIs 13.67 ± 0.58 – 29.50 ± 2.10 mm, by agar-well, and 11.33 ± 0.58 – 35.67 ± 2.52, by agar overlay, against human pathogenic bacteria, viz., Escherichia coli, Proteus vulgaris, Acinetobacter baumannii and Salmonella enterica serovar Typhi (Halder et al., 2017). The antibacterial activity of Lactobacillus fermentum LMEM 22 against gram-positive and gram-negative pathogenic bacteria, following agar-well diffusion method is depicted in Figure 3.
Gandevia et al. (2017) isolated, from cow milk, buffalo milk, goat milk and curd samples, a number of Lactobacillus species, including Lactobacillus fermentum, having the capacity to inhibit the growth of gram-positive bacteria, such as Staphylococcus aureus (ZDI: 8 – 17 mm) and Bacillus cereus (ZDI: 12 – 22 mm). The two Lactobacillus fermentum isolates from buffalo milk had ZDI of 19 mm, while the Lactobacillus oris conferred ZDI of 18 mm, against Listeria monocytogenes (Melia et al., 2017). The broad spectrum antibacterial activity of Lactobacillus fermentum has been demonstrated earlier (Ilayajara et al., 2011; Ramasamy and Suyambulingam, 2015; Podolsky, 1998), in which the LAB showed growth inhibitory activity against gram-positive (Staphylococcus aureus, Enterococcus spp., Streptococcus spp., Bacillus subtilis) and gram-negative (Proteus spp., Pseudomonas aeruginosa, Escherichia coli and Klebsiella pneumoniae) bacterial pathogens.
Kang et al. (2017) reported the enhancing killing of Staphylococcus aureus strains by Lactobacillus salivarius except Staphylococcus aureus ATCC 25923 strain, the growth of which was fully killed by Lactobacillus fermentum.
Sharma et al. (2016) isolated two lactic acid bacteria: Pediococcus acidilactici and Lactobacillus casei from milk cream and lassi, respectively, which had antibacterial activity against a large number of potential food-borne bacteria, viz., Staphylococcus aureus, Enterococcus faecalis, Listeria monocytogenes, Clostridium perfringens and Bacillus cereus having respective ZDIs range from 12 mm to 20 mm and from 13 mm to 19 mm. In the current study, the agar-well diffusion had ZDIs of 10.67±2.08 mm (Staphylococcus aureus) to 12±1.00 mm (Bacillus cereus), for gram-positive, and 13.00±2.65 mm (Escherichia coli) to 18.00±3.00 mm (Salmonella enterica Typhi), for gram-negative bacteria (Table 2). The Lactobacillus fermentum LMEM 22 isolate, following agar overlay method, also had antibacterial activity against the indicator bacterial strains (Figure 3), displaying ZDIs from 17.00±1.73 mm (Listeria monocytogenes) to 20.00±1.00 mm (Enterococcus faecalis), for gram-positive, and from 18.00±2.00 mm (Escherichia coli) to 33±2.65 mm (Acinetobacter baumannii) for gram-negative bacteria (Table 3).
| Table 1: Sugar fermentation test results for the isolated LAB from curd sample | |||
| Sugars | Utilization | Sugars | Utilization |
| Adonitol | – | D-Melezitose | + |
| Arabinose | W | Raffinose | + |
| Cellobiose | + | Rhamnose | + |
| Dextrose | + | D-Ribose | + |
| Esculin | – | Salicin | – |
| Glucose | + (g) | Sorbitol | + |
| Lactose | + | Sucrose | + |
| Mannitol | + | Trehalose | + |
| Mannose | + | Xylose | + |
| Melibiose | + | D-Galactose | + |
| +: Strong fermentation; w: week fermentation –: No fermentation | |||
The bacteriocin activity of the isolated LAB, Lactobacillus fermentum LMEM 22, has been shown in Table 2, while the Table 3 depicts the ‘R’ values of the isolated LAB. Iyapparaj et al. (2013) isolated Lactobacillus sp. MSU3IR strain, which against pathogenic bacteria: Staphylococcus aureus and Pseudomonas aeruginosa, displayed higher bacteriocin activity (393.2 ± 2.61 to 556.0 ± 5.34 AU/ml) in MRS medium, compared to the activity value (341.2 ± 2.36 to 473.2 ± 3.96 AU/ml) as recorded in Lactobacillus selection broth. Earlier, the ‘R’ values of curd lactobacilli strains ranged 3.00 – 13.17 mm, while the bacteriocin activity, in terms of arbitrary units (AU/ml), ranged 155.60 – 293.33, against MDR Klebsiella pneumoniae clinical isolates (Mandal and Halder, 2018).
| Table 2: Agar-well diffusion test results in terms ZDI (mm) and the calculated bacteriocin activity (Au/ml) of Lactobacillus fermentum LMEM22 against gram-positive and gram-negative indicator bacterial strains. | |||
| Indicator strains | Bacterial isolates | ZDI range (Mean ± SD) | Bacteriocin activity |
| Gram-negative | E. coli | 10-15 (13.00±2.65) | 173.33 |
| Pr. vulgaris | 12-14 (13.33±1.15) | 177.73 | |
| A. baumannii | 16-20 (17.33±2.31) | 231.07 | |
| Ps. aeruginosa | 15-16 (15.67±0.58) | 208.93 | |
| K. pneumoniae | 14-17 (15.67±1.53) | 208.93 | |
| S. enterica Typhi | 15-21 (18.00±3.00) | 240.00 | |
| Gram-positive | B. cereus | 11-13 (12.00±1.00) | 160.00 |
| E. faecalis | 10-13 (11.67±1.53) | 155.60 | |
| S. aureus | 9-13 (10.67±2.08) | 142.27 | |
| L. monocytogenes | 10-13 (11.67±1.53) | 155.60 | |
In another study, the probiotic lactobacilli had excellent antibacterial activity against gram-negative human pathogenic bacteria (Escherichia coli, Proteus vulgaris, Acinetobacter baumannii and Salmonella enterica serovar Typhi) displaying ‘R’ values ranging from 3.17 ± 0.29 to 15.33 ± 1.26 mm, and the bacteriocin activity ranging from 233.34 ± 45.54 to 280.56 ± 83.67 AU/ml (Halder et al., 2017). As per the report of Shehata et al. (2016), among nine isolates of LAB, one (Lactococcus lactic subsp. lactis) had strong activity (1600 AU/ml) against Klebsiella pneumoniae, while, four isolates had bacteriocin activity of 800 AU/ml against Escherichia coli (for Lactobacillus paracasei), Streptococcus pyogenes (for Lactobacillus gasseri), Staphylococcus aureus (for Lactobacillus rhamnosus) and Salmonella senftenberg (for Lactobacillus gasseri RM28). The current investigation demonstrates the capacity of antibacterial activity of Lactobacillus fermentum LMEM 22 for the indicator microorganisms, consisting of both gram-positive and gram-negative pathogenic bacteria, with an overall bacteriocin activity (AU/ml) of 142.27 – 240.00 and the ‘R’ values of 5.5 – 13.5 mm.
| Table 3: Agar overlay test results in terms ZDI (mm) and the calculated ‘R’ values for Lactobacillus fermentum LMEM22 against gram-positive and gram-negative indicator bacterial strains | |||
| Indicator strains | Bacterial isolates | ZDI range (Mean ± SD) | ‘R’ value (mm) |
| Gram-negative | E. coli | 16-20 (18.00±2.00) | 6.00 |
| Pr. vulgaris | 21-26 (23.33±2.52) | 8.67 | |
| A. baumannii | 31-36 (33.00±2.65) | 13.50 | |
| Ps. aeruginosa | 23-28 (26.00±2.65) | 10.00 | |
| K. pneumoniae | 20-25 (22.67±2.52) | 8.34 | |
| S. enterica Typhi | 20-25 (22.00±2.65) | 8.00 | |
| Gram-positive | B. cereus | 16-19 (17.67±1.53) | 5.84 |
| E. faecalis | 19-21 (20.00±1.00) | 7.00 | |
| S. aureus | 18-20 (18.67±1.56) | 6.34 | |
| L. monocytogenes | 15-18 (17.00±1.73) | 5.50 | |
| SD: standard deviation; ZDI: zone diameter of inhibition. | |||
The antibiotic susceptibility test results for Lactobacillus fermentum LMEM 22 isolate is shown in Figure 4. One of the most important probiotic features, defining safe for human consumption, of lactic acid bacteria is being their antibiotic sensitivity, and the intrinsic resistance (chromosomally conferred from point mutation) property as well (Georgieva et al., 2015). As has been demonstrated by Benavides et al. (2016), the isolated Lactobacillus fermentum was sensitive to ampicillin (ZDI: 28 mm), cefuroxime (ZDI: 30 mm), tetracycline (ZDI: 24 mm) and amoxicillin/clavulanic acid (ZDI: 26 mm) and resistant to gentamycin and kanamycin, and the LAB was found inhibitory to Escherichia coli (ZDI: 13 mm) and Salmonella Typhimurium (ZDI: 12 mm). As per our earlier report the curd lactobacilli had sensitivity to majority of the test antibiotics displaying a common resistance to Vm (Halder and Mandal, 2016). In the instant case, the isolated LAB: Lactobacillus fermentum LMEM 22 showed resistance to Ak, Cp, Km, Me and Vm (ZDI: ≤15 mm; range: 6 – 15 mm), and such resistances are intrinsic as well as non-transferable (Bamidele et al., 2017; Imperial and Ibana, 2016).
The Lactobacillus fermentum LMEM 22 was sensitive to Ac, Am, Cc, Cx, Gm, Im, Mp and Tc (ZDI: ≥21 mm; range: 22 – 34 mm), and intermediately susceptible to Cx and Tm with ZDIs of 16 and 18 mm (ZDI criteria range: 16 – 20 mm); the LAB, while, showed resistance (ZDI: ≤15 mm) to Ak, Cp, Km, Me and Vm. Thus, the isolated LAB, in this study, has been found to be safe, on the basis of lack of transferable antibiotic resistance property (Ammor et al., 2008; Imperial and Ibana, 2016), and this LAB might be useful in single-strain based probiotic formulation benefiting a large number of local population, in this part of the globe. It has been reported that multi-strain/multi-general probiotics might exhibit limited functional property for universal usage, requiring probiotics alternatives development and/or personalized probiotic approaches (Zmora et al., 2018; Suez et al., 2018).
Conclusion
The Lactobacillus fermentum LMEM22, which was isolated from locally available commercial curd, showed antagonistic activity against gram-positive as well as gram-negative pathogenic bacteria, with overall bacteriocin activity (AU/ml) of 142.27 – 240.00, and the ‘R’ value of 5.5 – 13.5, and (based upon the report available in literatures, too) there is no risk of transferable antibiotic resistance in the LAB. Thus, the isolated LAB might be useful as broad spectrum antibacterial biotherapeutics, and such native LAB isolate might be consumed alone, in place of antibiotic therapy, or can be used (based upon the antibiogram of the native LAB) in probiotic-antibiotic combination therapy. However, further studies are needed to validate the probiotic attributes of the isolated LAB, including its molecular identity as well as the antibiotic resistance management.
References
Ammor, M.S., Florez, A.B., van Hoek, A.H., de los Reyes-Gavilan, C.G., Aarts, H.J., Margolles, A and Mayo, B. (2008). Molecular characterization of intrinsic and acquired antibiotic resistance in lactic acid bacteria and bifidobacteria. Journal of Molecular Microbiology and Biotechnology, 14:6–15.
Bamidele, T.A., Adeniyi, B.A and Fowora, M.J. (2017). Antibiotic resistance patterns of lactic acid bacteria isolated from Nigerian grown salad vegetables. African Journal of Microbiology Research, 11: 433-439.
Bauer, A.J., Kirby, W and Turck, M. (1996). Antibiotic susceptibility testing by standardized single disc method. American Journal of Clinical Pathology, 45: 493–496.
Benavides, A.B., Ulcuango, M., Yepez, L and Tenea, G.N. (2016). Assessment of the in vitro bioactive properties of lactic acid bacteria isolated from native ecological niches of Ecuador. Revista Argentina De Microbiologia, 48:236-244.
Carasi, P., Diaz, M., Racedo, S.M., Antoni, G.D., Urdaci, M.C and Serradell, M.A. (2014). Safety characterization and antimicrobial properties of kefir-isolated Lactobacillus kefiri. Biomed Research International, 2: 1–7.
FAO/WHO (2001). Health and nutritional properties of probiotics in food including powder milk with live lactic acid bacteria: report of a Joint FAO/WHO expert consultation on evaluation of health and nutritional properties of probiotics in food including powder milk with live lactic acid bacteria, Technical Report, Food and Agriculture Organization/World Health Organization, Cordoba, Argentina. http://www.who.int/foodsafety/publications/fs management/en/probiotics.pdf.
FAO and WHO, 2007. WHO working group on drafting guidelines for the evaluation of probiotics in food. Guidelines for the evaluation of probiotics in food: Report of a joint FAO/WHO. FAO and WHO, London, Ontario, Canada.
Gandevia, H., Rana, N and Desai, B. (2017). Screening, production and antibacterial activity of bacteriocin from Lactobacillus spp. BMR Microbiology, 3: 1-8.
Georgieva, R., Yocheva, L., Tserovska, L., Zhelezova, G., Stefanova, N and Atanasova, A. (2015). Antimicrobial activity and antibiotic susceptibility of Lactobacillus and Bifidobacterium spp. intended for use as starter and probiotic cultures. Biotechnology and Biotechnological Equipment, 29: 84–91.
Halder, D and Mandal, S. (2015). Curd lactobacilli with probiotic potentiality. Translational Biomedicine, 6:1-6.
Halder, D and Mandal, S. (2016). Antibacterial potentiality of commercially available probiotic lactobacilli and curd lactobacilli strains, alone and in combination, against human pathogenic bacteria. Translational Biomedicine, 7: 1–7.
Halder, D., Mandal, M., Chatterjee, S.S., Pal, N.K and Mandal, S. (2017). Indigenous probiotic lactobacillus isolates presenting antibiotic like activity against human pathogenic bacteria. Biomedicines, 5: 1-11.
Mandal, S and Halder, D. (2018). Exploring Anti-Klebsiella pneumoniae activity of probiotic lactobacilli of curd origin. Acta Scientific Microbiology, 1: 49-53.
Holt, J.G and Krieg, N.R. (1984). Bergey’s Manual of Systematic Bacteriology; Williams and Wilkins: Baltimore, MD, USA.
Ilayajara, R., Radhamadhavan, P.A and Nirmala, C.B. (2011). Assessment for potential use of bacteriocin producing Lactobacillus fermentum isolated from human milk for preventing urinary tract infections. Journal of Pharmacy Research, 4: 3445–3447.
Imperial, I.C.V.J and Ibana, J.A. (2016). Addressing the antibiotic resistance problem with probiotics: reducing the risk of its double-edged sword effect. Frontiers in Microbiology, 7: 1-10.
Iyapparaj, P., Maruthiah, T., Ramasubburayan, R., Prakash, S., Kumar, C., Immanuel, G and Palavesam, A. (2013). Optimization of bacteriocin production by Lactobacillus sp. MSU3IR against shrimp bacterial Pathogens. Aquatic Biosystems, 9:1-10.
Kang, M.S., Lim, H.S., Jong-Suk Oh, J.K., Lim, Y.J., Wuertz-Kozak, K., Harro, J.M., Mark E. Shirtliff, M.E and Achermann, Y. (2017). Antimicrobial activity of Lactobacillus salivarius and Lactobacillus fermentum against Staphylococcus aureus. Pathogens and Disease, 75: 1-10.
Liasi, S.A., Azmi, T.I., Hassan, M.D., Shuhaimi, M and Rosfarizan, M. (2009). Antimicrobial activity and antibiotic sensitivity of three isolates of lactic acid bacteria from fermented fish product Budu. Malaysian Journal of Microbiology, 5: 33–37.
Mahalot, A and Mandal, S. (2018). Assessment of lactic acid bacteria from cow milk and goat milk samples for probiotic potentiality by in vitro methods. Acta Scientific Pharmaceutical Sciences, 2: 56-58.
Melia, S., Purwati, E., Yuherman, Jaswandi, Aritonang, S.N and Silaen, M. (2017). Characterization of the antimicrobial activity of lactic acid bacteria isolated from buffalo milk in west Sumatera (Indonesia) against Listeria monocytogenes. Pakistan Journal of Nutrition, 16: 645-650.
Nigam, A., Kumar, A., Madhusudan, H.V and Bhola, N. (2012). In-vitro screening of antibacterial activity of lactic acid bacteria against common enteric pathogens. Journal of Biomedical Sciences, 1: 1-6.
Nivien, A., Ghani, S.A.E., Gomaa, R.S and Fouad, M.T. (2016). Molecular identification of potential probiotic lactic acid bacteria strains isolated from Egyptian traditional fermented dairy products. Biotechnology, 15: 35-43.
Pisano, M.B., Viale, S., Conti, S., Fadda, M., Deplano, M., Melis, M.P., Deiana, M and Cosentino, S. (2014). Preliminary evaluation of probiotic properties of Lactobacillus strains isolated from Sardinian dairy products. Biomed Research International, 2: 1–8.
Podolsky, S. (1998). Cultural divergence: Elie Metchnikoff’s Bacillus bulgaricus therapy and his underlying concept of health. Bulletin of the History of Medicine, 72: 1–27.
Ramasamy, T.K and Suyambulingam, K. (2015). Molecular characterization of Lactobacillus sp. from Indian curd and its antagonistic effects on uropathogens of diabetic patients. International Research Journal of Biological Sciences, 4: 12–22.
Sharma, K., Sharma, N and Sharma, R. (2016). Identification and evaluation of in vitro probiotic attributes of novel and potential strains of lactic acid bacteria isolated from traditional dairy products of north-west Himalayas. Journal of Clinical Microbiology and Biochemical Technology, 2: 018-025.
Shehata, M.G., Sohaimy S.A.E., Malak, A and Youssef, E.M.M. (2016). Screening of isolated potential probiotic lactic acid bacteria for cholesterol lowering property and bile salt hydrolase activity. Annals of Agricultural Science, 61: 65–75.
Shokryazdan, P., Sieo, C.C., Kalavathy, R., Liang, J.B., Alitheen, N.B., Jahromi, M.F and Ahmed, M. (2014). Probiotic potential of Lactobacillus strains with antimicrobial activity against some human pathogenic strains. BioMed Research International, 2: 1–16.
Singh, B., Mal, G and Marotta, F. (2017). Designer probiotics: paving the way to living therapeutics. Trends in Biotechnology, 35: 679-681.
Suez, J., Zmora, N., Zilberman-Schapira, G., Mor, U., Dori-Bachash, M., Bashiardes, S., Zur, M., Regev-Lehavi, D., Brik, R.B.-Z., Federici, S., et al. (2018). Post-antibiotic gut mucosal microbiome reconstitution is impaired by probiotics and improved by autologous FMT. Cell 174, 1406–1423.
Tagg, J.R and McGiven, A.R. (1971). Assay system for bacteriocins. Applied Microbiology, 21: 943–944.
Van Boeckel, T.P., Gandra, S., Ashok, A., Caudron, Q., Grenfell, B.T., Levin, S.A and Laxminarayan, R. (2014). Global antibiotic consumption 2000 to 2010: an analysis of national pharmaceutical sales data. Lancet Infectious Disease, 14: 742–750.
Vlkova, E., Rada, V., Popelarova, P., Trojanová, I and Killer, J. (2006). Antimicrobial susceptibility of bifidobacteria isolated from gastrointestinal tract of calves. Livestock Science, 105: 253–259.
Vuotto, C., Longo, F and Donelli, G. (2014). Probiotics to counteract biofilm-associated infections: promising and conflicting data. International Journal of Oral Science, 6: 189–194.
Zmora, N., Zilberman-Schapira, G., Suez, J., Mor, U., Dori-Bachash, M., Bashiardes, S., Kotler, E., Zur, M., Regev-Lehavi, D., Brik, R.B.-Z., et al. (2018). Personalized gut mucosal colonization resistance to empiric probiotics is associated with unique host and microbiome features. Cell 174, 1388–1405.