Department of Biotechnology, Rama University, Kanpur, India.
Corresponding author email: viveksrivastavabio@gmail.com
Article Publishing History
Received: 26/04/2021
Accepted After Revision: 29/06/2021
Malaria parasite resistance to currently available drugs has emerged as a major global health problem. As a result, new approaches to developing more target-specific antimalarial drugs are urgently needed. In apicoplast, the fatty acid biosynthesis enzyme enoyl-acyl carrier protein reductase (FabI) plays a vital role in the growth of malaria parasite Plasmodium falciparum in the liver-stage; hence it is an excellent target for the development of novel antimalarials drugs. In order to identify novel inhibitors of the FabI enzyme, a computational investigation of the fatty acid production pathway in Plasmodium falciparum is presented. Latest research findings has shown that antimicrobial agent triclosan inhibits the growth of Plasmodium falciparum by inhibiting the enzyme Fab I. Virtual screening focused on ligand was used to identify triclosan analogs against drug libraries, bioactive, commercial and virtual compounds for inhibiting PfFabI with Swiss similarity tool.
Using SwissADME tool, screened compounds were subjected to physicochemical, pharmacokinetic, and toxicological analysis. Compound fulfilling ADME/Tox parameters is docked using MTiAutoDock server with binding pocket of FabI enzyme. The docked complexes were validated and enumerated based on the AutoDock scoring function to pick the best conformation. Triclosan analogs Pubchem CID 448623 and 71579715 having lowest binding energy with FabI enzyme even lower than known antimicrobial agent triclosan were predicted as novel inhibitors of PfFabI. To maximize accuracy of result, best predicted compounds again docked with FabI enzyme with different algorithm using PatchDock server. Our research has led to the discovery of two novel FabI inhibitors, which may be confirmed by laboratory experiments.
Enoyl-ACP Reductase, Fabi, Malaria, Plasmodium Falciparum, Triclosan.
Singh P, Srivastava V. In-silico identification of triclosan analogs as novel inhibitors of enoyl-ACP reductase from Plasmodium falciparum. Biosc.Biotech.Res.Comm. 2021;14(2).
Singh P, Srivastava V. In-silico identification of Triclosan analogs as Novel Inhibitors of Enoyl-ACP Reductase from Plasmodium falciparum. Biosc.Biotech.Res.Comm. 2021;14(2). Available from: <ahref=”https://bit.ly/3ioVlaR“>https://bit.ly/3ioVlaR</a>
Copyright © Singh and Srivastava This is an open access article distributed under the terms of the Creative Commons Attribution License (CC-BY) https://creativecommns.org/licenses/by/4.0/, which permits unrestricted use distribution and reproduction in any medium, provide the original author and source are credited.
INTRODUCTION
Malaria is a major public health issue that has become increasingly critical and continues to promote study, impacting the world’s main tropical and subtropical areas. 228 million malaria cases occurred worldwide in 2018 compared to 231 million in 2017 (WHO 2019). India and 19 countries in sub-Saharan Africa accounted for approximately 85% of the global malaria burden. An estimated 405 000 deaths from malaria were reported globally in 2018, compared with an estimated 416 000 deaths in 2017. Plasmodium falciparum has to be the most dominant malaria parasite in the South East Asia region of the WHO, accounting for 50% of confirmed cases of malaria in 2018 (WHO 2019).
Anti-malarial drug formulation; vaccination and vector control are the main methods to control this parasitic disease. Parasitic drugs are between these the first line of defence. The P. falciparum ‘chloroquine resistance transporter’ (PfCRT) was first identified as a main determinant of chloroquine (CQ) resistance, but mutations in this protein are now known to affect the parasite’s susceptibility to a variety of current and potential antimalarials (Martin, 2020). Thus, developing a new generation of anti-malarial drugs will represent a significant advance to consider. Apicoplast in malaria parasites has important consequences in combating their anti-malarial drug resistance (Gleeson, 2000; Liting and Geoffrey 2010).
Apicoplast metabolic pathways of isoprenoid precursor synthesis, fatty acid synthesis, heme synthesis and biogenesis of the iron sulfur cluster acting role in genome replication, transcription, translation, post-translational modification and protein turnover (Ralph et al. 2004; Liting and Geoffrey 2010). The apicoplast is a remnant of an ancient cyanobacterium that incorporates enzymes and biosynthetic processes that are more bacterial than eukaryotic in nature, allowing antimalarial agents to be designed that are selective against apicoplast targets (Ralph et al. 2004; Liting and Geoffrey 2010).
Apicoplast pathways fatty acid and isoprenoid precursor synthesis are clearly bacterial in origin and are potential targets for antibiotic drugs because they differ from the eukaryotic host processes.The Plasmodium FAS-II pathway was of particular therapeutic significance as it is distinct from the mammalian type I (FAS-I) pathway. The fatty acid biosynthesis enzyme enoyl-ACP reductase (FabI) plays a vital role in the growth of parasites in the liver-stage (Yu et al. 2008). Among the many known PfFabI inhibitors studied so far, triclosan remains the most active with an IC50 = 14 ng / ml (50 nM), but due to human health and environmental concerns, the compound is not appropriate for medicinal application (Kapoor et al. 2004). In this study, we have incorporated ligand based virtual screening method to search out the potential triclosan analogs for the treatment against maleria targeting the plasmodium falciparum enoyl-acyl-carrier-protein reductase, which were not reported previously.
MATERIAL AND METHODS
Ligand based virtual screening of triclosan against databases using Swisssimilarity web tool. Swisssimilarity tool’s screenable libraries include drugs, bioactive and commercial compounds and millions of virtual compounds readily synthesizable from commercially available synthetic reagents (Zoete et al. 2016).A drug with good oral absorption must meet the following parameters: molecular weight of less than 500 Da, logP (lipophilicity) of less than five (5); maximum of five ( 5) groups of hydrogen donors and maximum of ten (10) groups of accepters binding intestinal permeability and consisting of the first steps towards good oral bioavailability. In-silico physicochemical, pharmacokinetic and toxicological properties of triclosan analogs were perfomed by SwissADME web tool. The crystal structure of plasmodium falciparum enoyl-acyl-carrier-protein reductase with triclosan (PDB ID: 1NHG) was retrieved from the Protein Data Bank (https://www.rcsb.org/). All the Nicotinamide-adenine-dinucleotide (NAD) and triclosan compound were removed and polar hydrogen added to make the structure of enoyl-acyl-carrier-protein reductase prepared for molecular docking processes (Daina et al. 2017).
Binding site residues of FabI enzyme was predicted by CASTp3.0 server (Tian et al. 2018). Computed Atlas of Surface Topography of proteins (CASTp) is a web server that provides online resources to find delineate and quantify the geometric and topological properties of protein structures. The molecular docking method can be used to model the atomic level interaction between a small molecule and a protein, which enables us to describe the actions of small molecules in the target protein binding site as well as to elucidate basic biochemical processes. Screened triclosan analogs were docked with enoyl-ACP reductase using MTiAutoDock (https://mobyle.rpbs.univ-paris-diderot.fr/cgi-bin/portal.py#forms::MTiAutoDock). The most negative binding affinity score of compounds were chosen as candidate compounds (Daina et al. 2017).
RESULTS AND DISCUSSION
In silico ADME/Tox screening of triclosan analogs: About half of the drug candidates struggled during development due to ADME/Tox deficiencies. In order to prevent this development failure a series of In-silico ADME/Tox screens was introduced with the intention of discarding compounds during the discovery process. The ADME predictions of triclosan analogs for passive human gastrointestinal absorption (GI) and permeation of the blood-brain barrier (BBB) are both based on the BOILED-Egg model were shown in table 1. The properties of human intestinal absorption are determinant of the drug production and are required to be administered orally. The blood-brain barrier (BBB) plays a significant role in drug pharmacology (Ma et al. 2005).
Carcinogenicity is a cancer-causing risk in the body. Triclosan analogs were accessed by five different rule-based filters such as Lipinski filter implemented rule-of-five, Ghose, Veber, Egan and Muegge methods, respectively and shown in table 1. The result presented in Table 1 shows that all of the investigated compounds present high gastrointestinal absorption, good skin permeation and inhibit xenobiotic metabolism involving cytochrome CYP1A and CYP2D6.At the end of the entire ADMET evaluation screening, four compounds, CID 15624, 16220130, 25023959 and 156913, failed the drug likeliness test and were deleted for subsequent analysis (Ghose et al. 1999; Egan et al. 2000; Muegge et al. 2001; Veber et al. 2002; Lipinski 2004; Ma et al. 2005).
Table 1. The pharmacokinetics and drug likeliness properties of triclosan analogs.
| Sl.
No. |
Pubchem CID | GI | BBB | CYP1A2 inhibitor | CYP2D6 inhibitor | LogKp (Cm/s) | Bioavaliab-ility Score | Lipinski | Ghose | Veber | Egan | Muegge |
| 1 | 5564
(Triclosan) |
High | Yes | Yes | No | -4.69
|
0.56 | Yes | Yes | Yes | Yes | Yes |
| 2 | 7638
|
High | Yes | Yes | Yes | -5.09
|
0.55 | Yes | Yes | Yes | Yes | Yes |
| 3 | 18807 | High | Yes | Yes | No | -4.63
|
0.56 | Yes | Yes | Yes | Yes | Yes |
| 4 | 44129612 | High | Yes | Yes | No | -5.58 | 0.55 | Yes | Yes | Yes | Yes | Yes |
| 5 | 448978 | High | Yes | Yes | No | -5.04
|
0.56 | Yes | Yes | Yes | Yes | Yes |
| 6 | 447966 | High | Yes | Yes | Yes | -6.02 | 0.55 | Yes | Yes | Yes | Yes | Yes |
| 7 | 2255489
|
High | No | Yes | No | -6.74 | 0.55 | Yes | Yes | Yes | Yes | Yes |
| 8 | 448623 | High | Yes | Yes | Yes | -5.47
|
0.55 | Yes | Yes | Yes | Yes | Yes |
| 9 | 69545180 | High | Yes | Yes | Yes | -6.35
|
0.55 | Yes | Yes | Yes | Yes | Yes |
| 10 | 15624 | High | Yes | Yes | No | -5.17
|
0.56 | Yes | No | Yes | Yes | No |
| 11 | 16122582 | High | No | No | No | -6.95
|
0.55 | Yes | Yes | Yes | Yes | Yes |
| 12 | 16220130 | High | Yes | Yes | Yes | -4.69 | 0.55 | Yes | Yes | Yes | Yes | No |
| 13 | 3758 | High | No | No | No | -6.62
|
0.55 | Yes | Yes | Yes | Yes | Yes |
| 14 | 5564 | High | Yes | Yes | No | -4.69
|
0.55 | Yes | Yes | Yes | Yes | Yes |
| 15 | 18807
|
High | Yes | Yes | No | -4.63
|
0.55 | Yes | Yes | Yes | Yes | Yes |
| 16 | 5271320 | High | Yes | Yes | No | -4.84
|
0.55 | Yes | Yes | Yes | Yes | Yes |
| 17 | 23656591
|
High | Yes | Yes | Yes | -4.94
|
0.55 | Yes | Yes | Yes | Yes | Yes |
| 18 | 852148 | High | Yes | Yes | No | -5.50
|
0.55 | Yes | Yes | Yes | Yes | Yes |
| 19 | 45490026
|
High | Yes | No | No | -5.50
|
0.55 | Yes | Yes | Yes | Yes | Yes |
| 20 | 22947105
|
High | Yes | Yes | No | -4.75
|
0.55 | Yes | Yes | Yes | Yes | Yes |
| 21 | 71718966 | High | Yes | Yes | Yes | -4.42
|
0.55 | Yes | Yes | Yes | Yes | Yes |
| 22 | 44410251 | High | Yes | Yes | No | -5.74 | 0.55 | Yes | Yes | Yes | Yes | Yes |
| 23 | 25023958 | High | Yes | Yes | Yes | -4.53 | 0.55 | Yes | Yes | Yes | Yes | Yes |
| 24 | 44405311 | High | Yes | Yes | No | -5.40 | 0.55 | Yes | Yes | Yes | Yes | Yes |
| 25 | 21272512 | High | Yes | Yes | No | -5.27
|
0.55 | Yes | Yes | Yes | Yes | Yes |
| 26 | 11659169 | High | Yes | Yes | No | -5.28 | 0.55 | Yes | Yes | Yes | Yes | Yes |
| 27 | 6852148 | High | Yes | Yes | No | -5.50 | 0.55 | Yes | Yes | Yes | Yes | Yes |
| 28 | 25023955 | High | Yes | Yes | No | -5.28 | 0.55 | Yes | Yes | Yes | Yes | Yes |
| 29 | 71579715 | High | Yes | Yes | Yes | -5.10
|
0.55 | Yes | Yes | Yes | Yes | Yes |
| 30 | 25023959 | High | Yes | Yes | Yes | -4.23
|
0.56 | Yes | Yes | Yes | Yes | No |
| 31 | 5564 | High | Yes | Yes | No | -4.69 | 0.55 | Yes | Yes | Yes | Yes | Yes |
| 32 | 94509
|
High | Yes | Yes | No | -4.63 | 0.55 | Yes | Yes | Yes | Yes | Yes |
| 33 | 12774295
|
High | Yes | Yes | No | -4.70 | 0.55 | Yes | Yes | Yes | Yes | Yes |
| 34 | 10846712 | High | Yes | Yes | No | -4.97 | 0.55 | Yes | Yes | Yes | Yes | Yes |
| 35 | 10711808
|
High | Yes | Yes | No | -5.45 | 0.55 | Yes | Yes | Yes | Yes | Yes |
| 36 | 82267072
|
High | Yes | Yes | No | -5.10
|
0.55 | Yes | Yes | Yes | Yes | Yes |
| 37 | 21272514 | High | Yes | Yes | No | -4.53 | 0.55 | Yes | Yes | Yes | Yes | Yes |
| 38 | 12346120
|
High | Yes | Yes | No | -4.61 | 0.55 | Yes | Yes | Yes | Yes | Yes |
| 39 | 82267104 | High | Yes | Yes | Yes | -5.03 | 0.55 | Yes | Yes | Yes | Yes | Yes |
| 40 | 82053313 | High | Yes | Yes | No | -5.39 cm/s
|
0.55 | Yes | Yes | Yes | Yes | Yes |
| 41 | 14094464 | High | Yes | No | No | -5.66
|
0.55 | Yes | Yes | Yes | Yes | Yes |
| 42 | 45093134
|
High | Yes | No | No | -5.59
|
0.55 | Yes | Yes | Yes | Yes | Yes |
| 43 | 59319916
|
High | Yes | Yes | Yes | -5.04
|
0.55 | Yes | Yes | Yes | Yes | Yes |
| 44 | 156913 | High | No | No | No | -4.22
|
0.56 | Yes | No | Yes | No | No |
| 45 | 627458 | High | Yes | No | No | -4.52 | 0.55 | Yes | Yes | Yes | Yes | Yes |
GI-Gastrointestinal absorption, BBB-Blood Brain Barrier penetration,CYP: Cytochrome P450, Log Kp-Skin Permeation Coefficient.
Binding site analysis of FabI: CASTp(http://sts.bioe.uic.edu/castp/calculation.html) provides a comprehensive and detailed quantitative characterization of the inner voids and surface pockets on FabI’s three-dimensional structure. Identified binding site residues of Fab I on chain A of Plasmodium FalciparumareGLY104, ILE105, GLY106, ASP107, GLY110, TYR111, GLY112, TRP113, GLY129, TRP131, VAL134, PHE167, ASP168, ALA169, SER170, HIS214, SER215, LEU216, ALA217, ASN218, ALA219, VAL222, LYS240, SER241, LEU265, THR266, TYR267, TYR277, MET281, LYS285, LEU288, ALA312, GLY313, PRO314, LEU315, SER317, ARG318, ALA319, ALA320, ALA322, ILE323.
Analysis of Molecular docking: Forty oneADME/Tox parameters qualify triclosan were docked in binding site of FabI enzyme using MTiAutoDock server.Binding energy of triclosan analogs were shown in table 2. On analysis of binding energy of analogs, it was found that two analogs, CID 448623 and 71579715 had low binding affinity -5.10 kcal / mol, -5.05 kcal / mol with FabI, respectively, even lower than -4.36 kcal / mol with triclosan, highlighted with bold in table 1.To maximize accuracy of result, best predicted analogs again docked with FabI enzyme from different algorithm using PatchDock server using PatchDockserver(Duhovny et al. 2002; Schneidman-Duhovny et al. 2005). The clustering RMSD was a default value of 4.0. The top 10 PatchDock solutions produced were refined using Firedock based on their binding energyand the best models had global energy of -39.04 and -43.33kcal/mol for analogs CID 448623 and 71579715 that suggests a good interaction between ligand and protein (Andrusier et al. 2007; Mashiach et al. 2008).
Table 2. Triclosan analogs physicochemical properties and binding energy with FabI
| Sl.No. | Pubchem CID | Molecular weight (g/mol) | No of rotatable bonds | No of H-bond acceptors | No of H-bond donors | TPS (Ų) | Binding energy (kcal/mol) |
| 1 | 5564(Triclosan)
|
289.5 | 2 | 2 | 1 | 29.5 | -4.36 |
| 2 | 7638
|
200.23 | 3 | 2 | 1 | 29.5 | -4.76 |
| 3 | 18807 | 255.09 | 2 | 2 | 1 | 29.5 | -4.08 |
| 4 | 44129612 | 384.7 | 2 | 3 | 1 | 65.8 | -4.42 |
| 5 | 448978
|
327.2 | 5 | 4 | 1 | 55.8
55.8 55.8 55.8 |
-4.42 |
| 6 | 447966 | 272.3 | 2 | 3 | 1 | 54.5 | -4.71 |
| 7 | 2255489 | 239.3 | 2 | 4 | 2 | 99.3 | -4.86 |
| 8 | 448623
|
338.17 | 3 | 3 | 2 | 49.3 | -5.10 |
| 9 | 69545180 | 240.26 | 1 | 2 | 3 | 65.1 | -3.60 |
| 10 | 16122582
|
257.31 | 6 | 5 | 1 | 94.8 | -3.28 |
| 11 | 3758
|
222.24 | 2 | 3 | 1 | 69.3 | -3.48 |
| 12 | 5564 | 289.5 | 2 | 2 | 1 | 29.5 | -4.71 |
| 13 | 18807
|
255.09 | 2 | 2 | 1 | 29.5 | -4.07 |
| 14 | 5271320 | 220.65 | 2 | 2 | 1 | 29.5 | -4.14 |
| 15 | 23656591
|
303.6 | 3 | 2 | 1 | 29.5 | -4.30 |
| 16 | 6852148
|
270.11 | 2 | 3 | 2 | 55.5 | -4.67 |
| 17 | 45490026 | 270.11 | 2 | 3 | 2 | 55.5
|
-3.34 |
| 18 | 22947105
|
269.12 | 2 | 2 | 1 | 29.5 | -4.45 |
| 19 | 71718966 | 270.71 | 2 | 2 | 1 | 29.5 | -4.94 |
| 20 | 44410251 | 235.66
|
2 | 3 | 2 | 55.5 | -4.45 |
| 21 | 25023958
|
283.1 | 3 | 2 | 1 | 29.5 | -4.24 |
| 22 | 44405311
|
294.1
|
3 | 3 | 1 | 53.2 | -4.52 |
| 23 | 21272512
|
271.09 | 2 | 3 | 2 | 49.7 | -4.49 |
| 24 | 11659169 | 280.1 | 2 | 3 | 1 | 53.2 | -4.72 |
| 25 | 6852148 | 270.11
|
2 | 3 | 2 | 55.5 | -4.66 |
| 26 | 25023955
|
280.1 | 2 | 3 | 1 | 53.2 | -4.66 |
| 27 | 71579715 | 271.7 | 2 | 3 | 1 | 42.2 | -5.05 |
| 28 | 5564 | 289.5 | 2 | 2 | 1 | 29.5 | -4.41 |
| 29 | 94509
|
255.09 | 2 | 2 | 1 | 29.5 | -3.78 |
| 30 | 12774295
|
220.65 | 2 | 2 | 1 | 29.5 | -4.12 |
| 31 | 10846712 | 222.67 | 1 | 2 | 1 | 29.5 | -3.34 |
| 32 | 10711808
|
334.54 | 1 | 2 | 1 | 29.5 | -3.60 |
| 33 | 82267072 | 221.08 | 2 | 2 | 1 | 29.5 | -3.39 |
| 34 | 21272514 | 255.09 | 2 | 2 | 1 | 29.5 | -3.61 |
| 35 | 12346120
|
220.65 | 2 | 2 | 1 | 29.5 | -3.39 |
| 36 | 82267104 | 221.08 | 2 | 2 | 1 | 29.5 | -4.27 |
| 37 | 82053313 | 186.63 | 1 | 2 | 1 | 29.5 | -3.68 |
| 38 | 14094464 | 253.08 | 3 | 4 | 1 | 47.9 | -3.01 |
| 39 | 45093134
|
209.02
|
3 | 3 | 2 | 49.7 | -3.18 |
| 40 | 59319916
|
200.66
|
3 | 2 | 1 | 29.5 | -3.39 |
| 41 | 627458
|
303.6 | 3 | 2 | 0 | 18.5 | -4.20 |
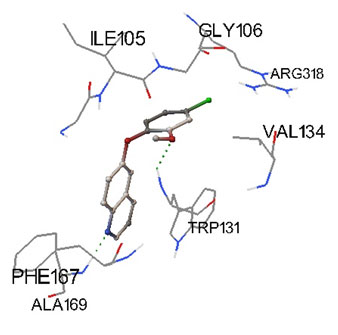
Predicted docked complexes were analyzed through Python Molecular Viewer for their interaction study shown in Figure 1 and 2. Triclosan analog was represented in sticks and balls model. Triclosan analog CID 448623 interacted with residues GLY104, LEU216, ASN218, ILE130, GLY129, TRP131, LYS240, SER170, ALA169, ASP168 and PHE167. All the residues interacting with CID 448623 belongs to predicted binding site residues of FabI except ILE130. Triclosan analog CID 71579715 interacted with residues GLY106, ARG316, ILE105, GLY106, VAL134, TRP131, ALA169, and PHE167. All the residues interacting with CID 71579715 also belongs to predicted binding site residues except ARG316. Interacting residues were represented in lines as Figure 1 and 2 (Sanner 1999; Pamudi et al. 2017).
Figure 1: Docking pose of triclosan analog CID448623in binding site of FabI enzyme.
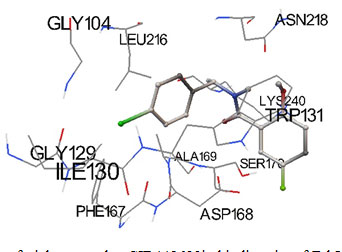
Figure 2: Docking pose of triclosan analog CID 71579715in binding site of FabI enzyme.Two H-bonds were formed between amino acid TRP131 and ALA169 of protein with compound, respectively. Hydrogen bonds are represented with spherical line.
The lack of an effective vaccine and the emergence of Plasmodium variants resistant to antimalarial agents emphasize the need for novel chemotherapeutic drugs. In-silico screening, also known as virtual screening, is currently being researched as a strategy for finding antimalarial medicines. Pamudi et al. (2017) explore at the usage of virtual screening to identify Enoyl Acyl Carrier Protein Reductase inhibitors in Plasmodium falciparum. The medicinal plants in Indonesia database were used in conjunction with a molecular docking method using GOLD software. They discovered two prospective inhibitor chemicals in tea that have the potential to be developed as antimalarial drugs: kaempferol 3-rhamnosyl-(1-3)-rhamnosyl-(1-6)-glucoside and epigallocatechin 3,5, -di-O-gallate (Pamudi et al. 2017; Dayse et al. 2020).
By using hierarchical virtual screening, Dayse et al. (2020) discovered flavonoids to be inhibitors of Plasmodium falciparum enoyl-acyl carrier protein (ACP) reductase (Dayse et al. 2020). A flavonoid library from the ChEMBL database was screened for physico-chemical similarity using the Euclidean distance as a criterion, and then molecular docking was performed using the GridScore scoring tool in the DOCK 6.5 software (Dayse et al. 2020). Using ligands from the Indonesian Medicinal Plants Database, in-silico screening was carried out using the AUTODOCK VINA software to identify potential Plasmodium falciparum enoyl-acyl carrier protein (ACP) reductase inhibitor candidates by Malau et al. (2020).
CONCLUSION
The current research utilises computer-based virtual screening to identify Plasmodium falciparum Enoyl-ACP reductase inhibitors that are required for malaria treatment. From several million chemical structures and a sequence of steps of rational refinement, including similarity search, ADME/Tox properties and molecular docking, we identified two triclosan analogs, CID 448623 and 71579715, as good inhibitors for further experimental testing.
ACKNOWLEDGEMENTS
Authors respectfully thanks the Department of Biotechnology, Rama University, Kanpur, Uttar Pradesh, India, for providing the essential computer facilities and consistent supervision during the research activity.
Conflict of interests: There are no conflicts of interest declared by the authors.
REFERENCES
Andrusier, N., Nussinov, R., and Wolfson, H.J. (2007). FireDock: Fast Interaction Refinement in Molecular Docking. Proteins, 69(1): 139-159.
Butina,D., Segall, M.D., and Frankcombe, K. (2002). Predicting ADME properties in silico: Methods and models. Drug Discov.Today, 7: S83–S88.
Daina, A., Michielin, O., and Zoete, V. (2017). SwissADME: A free web tool to evaluate pharmacokinetics, drug-likeness and medicinal chemistry friendliness of small molecules. Sci. Rep., 7:42717.
Dayse, A. A. S., Diego, M. da Costa, Larissa, M. O., Hugo, N. B., Clayton, Q. A., Aníbal, F. S., and Manoelito, C. dos Santos Jr. (2020) Identification of Flavonoids as Inhibitors of Plasmodium falciparum Enoyl-ACP Reductase by Hierarchical Virtual Screening. J. Braz. Chem. Soc., 31(12): 2544-2552.
Duhovny, D., Nussinov, R., and Wolfson, H.J. (2002). Efficient Unbound Docking of Rigid Molecules. In Gusfield et al. Ed. Proceedings of the 2’nd Workshop on Algorithms in Bioinformatics (WABI) Rome, Italy, Lecture Notes in Computer Science, Springer Verlag , 2452:185-200.
Egan, W.J., Merz, K.M., and Baldwin, J.J. (2000). Prediction of Drug Absorption Using Multivariate Statistics. J. Med. Chem., 43, 3867–3877.
Ghose,A.K., Viswanadhan, V.N., and Wendoloski, J.J. (1999). A knowledge-based approach in designing combinatorial or medicinal chemistry libraries for drug discovery.1. A qualitative and quantitative characterization of known drug databases. J Comb. Chem., 1:55–68.
Gleeson, M.T. (2000). The plastid in Apicomplexa: what use is it? Int J Parasitol., 30(10):1053-70.
Kapoor, M., Reddy, C.C., Krishnasastry, M.V., Surolia, N., and Surolia, A. (2004). Slow-tight-binding inhibition of enoyl-acyl carrier protein reductase from Plasmodium falciparum by triclosan. Biochem J., 381:719–724.
Lipinski, C.A. (2004). Lead- and drug-like compounds: the rule-of-five revolution. Drug Discov Today Technol.,1(4):337–341.
Liting, L., and Geoffrey, I.M. (2010). The evolution, metabolism and functions of the apicoplast. Philos Trans R Soc Lond B Biol Sci., 365(1541): 749-63.
Ma X., Chen, C., and Yang, J. (2005). Predictive Model of Blood-Brain Barrier Penetration of Organic Compounds. Acta Pharmacologica Sinica, 26:500-512.
Mashiach, E., Schneidman-Duhovny,D., Andrusier,N., Nussinov,R., and Wolfson, H.J. (2008). FireDock: a web server for fast interaction refinement in molecular docking. Nucleic Acids Res., 36: W229-32.
Martin, R. E. (2020). The transportome of the malaria parasite. Biol. Rev. Camb. Philos. Soc.95:305–332.
Malau, N. D.,and Azzahra, S. F.(2020). Analysis Docking of Plasmodium falciparum Enoyl Acyl Carrier Protein Reductase (Pfenr) With Organic Compunds from Virtual Screening of Herbal Database. Acta. Chim. Asiana., 3(1):127–134.
Min, Y., Kumar T.R.S., Nkrumah, L.J., Coppi,A., and Retzlaff S. (2008). The Fatty Acid Biosynthesis Enzyme FabI Plays a Key Role in the Development of Liver-Stage Malarial Parasites. Cell Host and Microbe, 4:567–578.
Muegge, I., Heald, S.L., and Brittelli, D. (2001). Simple selection criteria for drug-like chemical matter. J. Med. Chem., 44:1841–1846.
Patrick, G. L. (2020). Chapter 9 – Drug targets in the apicoplast. Antimalarial Agents, Design and Mechanism of Action, 319-374.
Pamudi, B. F., Azizahwati, A., and Yanuar, A. (2017). In-silico screening against antimalarial target Plasmodium falciparum enoyl-acyl carrier protein reductase. Asian Journal of Pharmaceutical and Clinical Research, 10(17):127-9.
Ralph, S.A., van Dooren, G.G., Waller, R.F., Crawford, M.J., Fraunholz, M.J., and Foth, B.J., (2004). Tropical infectious diseases: metabolic maps and functions of the Plasmodium falciparum apicoplast. Nat Rev Microbiol., 2(3):203-16.
Sanner, M.F. (1999). Python: A Programming Language for Software Integration and Development. J Mol Graph Model 1999, 17(1):57-61.
Schneidman-Duhovny, D., Inbar, Y., Nussinov, R., and Wolfson, H.J. (2005). PatchDock and SymmDock: servers for rigid and symmetric docking. Nucl. Acids. Res., 33: W363-367.
Tian,W., Chen,C., Lei, X., Zhao, j., and Liang J.(2018).CASTp3.0: computed atlas of surface topography of proteins. Nucleic Acids Research, 46(W1): W363–W367.
Veber, D.F.,Johnson, S.R., Cheng, H., Smith, B.R., Ward, K.W., and Kopple, K.D. (2002). Molecular properties that influence the oral bioavailability of drug candidates. J. Med. Chem., 45:2615–2623.
WHO (2019). World malaria report 2019. Available at https://www.who.int/news-room/feature-stories/detail/world-malaria-report-2019.
Zoete, V., Daina, A., Bovigny, C., and Michielin, O. (2016). SwissSimilarity: A Web Tool for Low to Ultra High Throughput Ligand-Based Virtual Screening., J. Chem. Inf. Model., 56(8):1399-1404.


