1Government Medical College, Bhavnagar-364001, Gujarat, India
2Department of Biosciences, Veer Narmad South Gujarat University, Surat-395007, Gujarat, India
3C.U. Shah Medical College, Surendranagar-363001, Gujarat, India
Corresponding author email: kotharidhyey17@gmail.com
Article Publishing History
Received: 15/02/2025
Accepted After Revision: 25/06/2025
This study investigates the potential of phytochemical compounds as natural inhibitors of human α-amylase (PDB ID: 1B2Y) for the management of type 2 diabetes mellitus (T2DM). Molecular docking was performed on a library of 3,320 phytochemicals derived from 20 medicinal plants and synthetic anti-diabetic compounds. The top-ranked compounds exhibited superior binding affinities (-11.7 to -18.6 kcal/mol) compared to the control drug Acarbose (-8.7 kcal/mol). Goyaglycoside-g, Goyaglycoside-e, and Momordicoside R emerged as the most potent inhibitors, with Momordicoside R specifically targeting the catalytic residue TYR151. Compounds from Momordica charantia (Kuguacin M, β-Amyrin, α-Amyrin) and Azadirachta indica (Melianoninol, Odoratone) also showed remarkable inhibitory effects. Interaction analysis revealed diverse binding mechanisms, including van der Waals forces, hydrogen bonding, and hydrophobic interactions with key residues such as TYR151 and TRP58. ADME analysis highlighted the challenges of limited bioavailability for some glycosides, while toxicity predictions identified Melianoninol as a promising candidate with a balanced efficacy-safety profile.
This study validates the ethnopharmacological use of Momordica charantia and Azadirachta indica in T2DM management and emphasizes the potential of phytochemicals as safer alternatives to synthetic drugs. Future research should focus on structural optimization and advanced formulations to enhance the bioavailability of these compounds. The present analysis demonstrates the superior efficacy of phytochemicals as natural α-amylase inhibitors over synthetic drugs, with identified compounds from Momordica charantia and Azadirachta indica showing 2.14-fold better binding affinity than Acarbose, establishing a foundation for developing safer, bioavailable anti-diabetic therapeutics through computational-guided structural optimization and advanced formulation strategies for clinical translation in type 2 diabetes management.
Phytochemicals, Type 2 Diabetes, Molecular Docking, Momordica charantia, Azadirachta indica
Kothari D. R, Godhaniya M. D, Kikani K. M. In silico Evaluation of Plant-Derived Compounds as Potential α-Amylase Inhibitors for Treatment of Type 2 Diabetes. Biosc.Biotech.Res.Comm. 2025;18(2).
Kothari D. R, Godhaniya M. D, Kikani K. M. In silico Evaluation of Plant-Derived Compounds as Potential α-Amylase Inhibitors for Treatment of Type 2 Diabetes. Biosc.Biotech.Res.Comm. 2024;18(2). Available from: <a href=”https://shorturl.at/uLjue“>https://shorturl.at/uLjue</a>
INTRODUCTION
Diabetes mellitus, particularly type 2 diabetes mellitus (T2DM), poses a significant global health challenge due to its high prevalence and associated complications. The prevalence of diabetes mellitus among adults has notably increased, from 4.7% in 1980 to 8.5% in 2014, affecting over 422 million people globally by 2014 (Kanter and Bornfeldt, 2016). In 2017 alone, approximately 462 million individuals were affected by type 2 diabetes, accounting for about 6.28% of the world’s population (Khan et al., 2019).
This condition is now the ninth leading cause of mortality, with over 1 million deaths yearly attributed to diabetes causes alone. One Recent study found that gamma-mangostin interacts with the active site of the alpha-amylase enzyme, showing a binding affinity of -9.1 kcal/mol, which is comparable to the control, acarbose (-16.4 kcal/mol). The interactions involve hydrogen, alkyl, and van der Waals bonds. In vitro testing further supported its potential, with gamma-mangostin reducing blood sugar levels by 43.33% compared to acarbose’s 56.25% at the same concentration (Kurniawan, Marfu’ah and Fazriah 2025).
Type 2 diabetes is primarily characterized by insulin resistance and pancreatic beta-cell dysfunction, leading to chronic hyperglycemia (DeFronzo et al., 2015). Factors contributing to insulin resistance include impaired insulin signaling, increased hepatic glucose production, and low-grade systemic inflammation. Furthermore, mitochondrial dysfunction in pancreatic cells exacerbates insulin and glucagon secretion issues (Grubelnik et al., 2020). Insulin resistance is closely linked to lipid metabolism alterations, which manifest as low HDL cholesterol levels, contributing to cardiovascular risks associated with T2DM (Vollenweider, Von Eckardstein and Widmann, 2015).
Recent research has shown promise in the development of novel therapeutic compounds. For example, a study on certain pyridazine derivatives (Molecules 45, 46, and 29) found they had good docking interaction scores with alpha-amylase receptors. While ADMET analysis revealed some limitations, such as lower solubility and poor distribution, these molecules generally maintained drug-like properties with good oral bioavailability. Notably, Molecule 46 was identified as non-toxic. In vitro tests further confirmed their potent alpha-amylase inhibition, with significantly lower IC50 values (a measure of how much of a substance is needed to inhibit a given biological process by half) compared to the standard drug acarbose (Varshney et al., 2024).
Current therapeutic approaches for managing T2DM focus on lifestyle modifications and pharmacological interventions. First-line treatments typically involve lifestyle changes complemented with metformin, which helps improve glycemic control. Other medications include sulfonylureas, thiazolidinediones, alpha-glucosidase inhibitors, glucagon-like peptide-1 agonists, and insulin therapy. However, these treatments often have limitations such as side effects and limited effectiveness over time (El-Kaissi and Sherbeeni, 2011a). The compounding issue of side effects with oral hypoglycemic agents severely affects patient compliance. The development of novel therapeutics, such as nucleic acid-based therapies, aims to overcome these limitations by targeting the expression of genes that cause insulin resistance and hyperglycemia (Kokil et al., 2015).
Mesenchymal stem cells also present a promising therapeutic avenue due to their potential to facilitate beta-cell regeneration and immune system regulation, although the associated risks require further investigation. Efforts are continually being made to discover effective interventions that can better manage T2DM and its complications, and these efforts emphasize the complex nature of the disease and the need for multidisciplinary approaches to treatment (El-Kaissi and Sherbeeni, 2011a Mikłosz and Chabowski, 2023).
α-Amylase is a critical enzyme in carbohydrate digestion and glucose absorption, being instrumental in breaking down dietary starch into simpler sugars like maltose and maltotriose. The enzyme’s activity leads to the liberation of glucose, which subsequently influences blood glucose levels and can aggravate postprandial hyperglycemia, a significant concern in diabetes management (Kashtoh and Baek, 2023).
Given its role, α-amylase presents a promising therapeutic target for managing diabetes. The inhibition of α-amylase slows down carbohydrate digestion and delays glucose absorption, thus helping regulate blood glucose levels (Sales et al., 2012a). Numerous studies have focused on natural plant-based compounds to inhibit α-amylase effectively, offering an alternative to synthetic inhibitors like acarbose and miglitol, which are known for their side effects such as gastrointestinal discomfort. Recent Study found that chlorogenic acid exhibited the highest docking scores against both AMPD1 (-8.41 kcal/mole) and PKA (-12.56 kcal/mole), suggesting its potential as a potent antidiabetic compound (Devakrishna, Taj and Upadhyay, 2024).
Phytochemicals, bioactive compounds derived from plants, offer significant potential as natural therapeutics, especially in the context of diabetes management. These compounds, including flavonoids, terpenes, phenolic acids, and alkaloids, have been investigated for their health-promoting properties and potential to act as anti-diabetic agents (Silva et al., 2017a; Vinayagam, Xiao and Xu, 2017a).The primary advantages of plant-derived compounds include their natural origin, potential lower toxicity, and fewer side effects compared to synthetic drugs . These attributes make phytochemicals an appealing alternative in therapeutic interventions, particularly where synthetic medications have limited effectiveness or cause adverse side effects over long-term use (A. Omara et al., 2010). Additionally, phytochemicals can be incorporated into dietary regimes, thus providing a cost-effective strategy for disease prevention and management (Silva et al., 2017b).
Classes of Phytochemicals with Anti-Diabetic Properties: Different classes of phytochemicals possess anti-diabetic properties through varied mechanisms. For instance:
- Flavonoids and Terpenes: Known for their antioxidant and anti-inflammatory actions, these compounds can inhibit key enzymes like α-amylase and α-glucosidase that are involved in carbohydrate digestion, thereby reducing blood glucose levels (Vinayagam, Xiao and Xu, 2017b).
- Phenolic Compounds: These are abundant in edible plants and have been associated with the regulation of glucose metabolism and improvement of insulin sensitivity (Silva et al., 2017b).
- Alkaloids and Lignans: These compounds enhance insulin secretion and sensitivity, contribute to glycogen synthesis, and modulate oxidative stress, which are beneficial in diabetes management (Ardalani et al., 2021).
Molecular docking plays a pivotal role in drug development by simulating and predicting interactions between small molecules and target proteins, thereby streamlining the process of drug discovery (Mursal et al., 2024a). As a structure-based method, it aids in the identification of novel therapeutic compounds and the prediction of ligand-target interactions (Pinzi and Rastelli, 2019). Molecular docking is integral to virtual screening and lead optimization, where it expedites the identification of potential drug candidates by allowing high-throughput evaluation of molecular interactions. The software used in molecular docking, such as AutoDock and GOLD, utilizes algorithms to predict binding modes and assess binding affinities, significantly contributing to the drug development process (Mursal et al., 2024b).
Structure-based drug design (SBDD) relies heavily on the three-dimensional structure of target biomolecules to guide drug development (Schneuing et al., 2024). It involves the fitting of drug-like molecules into a protein’s binding site, enhancing the search for drug candidates through rational drug design strategies (Bentham Science Publisher, 2006). SBDD benefits from advanced computational techniques such as virtual screening and ensemble docking, which improve the precision of docking predictions and accelerate drug discovery (Mathur et al., 2024). Despite its advantages, SBDD faces challenges such as the need for accurate binding site identification and predicting the effects of protein-ligand interactions accurately.
While molecular docking and structure-based drug design offer significant advances in drug development, careful application and understanding of their limitations are necessary for their effective use in pharmaceutical research (Patel et al., 2022). In silico docking and ADMET studies on clinical targets for type 2 diabetes correlated to in vitro inhibition of pancreatic alpha-amylase and alpha-glucosidase by rutin, caffeic acid, p-coumaric acid, and vanillin,” published in 2025, investigate the potential of several natural compounds as inhibitors for Type 2 Diabetes. The study combines in silico (computational) docking and ADMET (Absorption, Distribution, Metabolism, Excretion, and Toxicity) analyses with in vitro (laboratory) inhibition studies of pancreatic alpha-amylase and alpha-glucosidase (McMillan, Bester and Apostolides, 2025).
α-Amylase plays a crucial role in carbohydrate digestion by catalyzing the hydrolysis of starch into sugars. The inhibition of α-amylase is considered a significant strategy for managing conditions like diabetes and obesity due to its role in reducing postprandial blood glucose levels (Sales et al., 2012a). The enzyme functions as a calcium metalloenzyme, facilitating the breakdown of polysaccharides, which subsequently leads to increased blood glucose levels (Kaur et al., 2021).
The α-amylase enzyme is a well-characterized protein with a detailed structural analysis underscoring its catalytic mechanisms. The crystal structure of α-amylase has been studied extensively using techniques like X-ray diffraction, providing insights into its active sites and functionality. The active site of α-amylase involves key amino acid residues that are essential for its catalytic action of hydrolyzing starch. The process of starch breakdown is initiated through the enzyme’s active site, where the polysaccharide chains are cleaved into monosaccharides and disaccharides. This enzyme’s action is central to carbohydrate metabolism, particularly in the digestion process (Jayaraj, Suresh and Kadeppagari, 2013a).
Multiple studies have explored the development of α-amylase inhibitors as therapeutic agents, particularly for diabetes management. These inhibitors function by binding to key sites on the enzyme, thereby preventing its interaction with substrates. Acarbose, a well-known α-amylase inhibitor, interacts with the enzyme’s active site, effectively reducing glucose production from carbohydrate digestion (Li et al., 2021). Additionally, novel inhibitors like those based on the pyrazole motif have shown promising potential due to their strong binding affinities and effective inhibitory actions on α-amylase.
Research into plant-based inhibitors has identified several compounds, particularly flavonoids, that exhibit significant α-amylase inhibitory activities. The efficacy of these inhibitors is often linked to their molecular structures, such as the number of hydroxyl groups, which enhance their binding affinity to the enzyme (Sales et al., 2012a; Ayorinde et al., 2025). Flavonoids, in particular, have been highlighted for their dual inhibition capabilities against α-amylase and α-glucosidase, making them promising candidates for natural antidiabetic therapies(Ayorinde et al., 2025).
Phytochemical inhibitors, particularly those targeting the enzyme α-amylase, hold promise as natural treatments for diabetes and related metabolic disorders. Recent studies have focused on identifying and validating such inhibitors derived from plant sources, exploring their structure-activity relationships (SAR), and experimentally validating their efficacy. The primary objective of this study is to screen phytochemicals against the 1B2Y protein target, which is likely an α-amylase enzyme. The secondary objectives involve analyzing the binding modes of these phytochemicals with 1B2Y and predicting their ADMET (Absorption, Distribution, Metabolism, Excretion, and Toxicity) properties. The hypothesis suggests that natural compounds have the potential to effectively inhibit α-amylase activity, which could have implications for managing conditions related to carbohydrate metabolism.
METHODOLOGY
The Phytochemical Library creation: An extensive literature review was conducted to investigate the anti-diabetic properties of plants using an in-silico method, leading to the selection of bioactive plants. The selected plants were analyzed using the KNApSAcK: A Comprehensive Species-Metabolite Relationship (Afendi et al., 2012) and IMPPAT: Indian Medicinal Plants, Phytochemistry, and Therapeutics databases (Mohanraj et al., 2018), from which phytochemicals linked to different plants were collected. The assembled list of phytochemicals was subsequently organized, and their 3D structural representations were acquired in SMILE format from the PubChem database (Kim et al., 2025).
The target alpha-amylase main protease preparation: The crystal structures of the alpha-amylase (PDB ID: 1B2Y, 3.20 Å) were obtained from the protein data bank (https://www.rcsb.org). The selected protein structures were downloaded along with its co-crystallized Acarbose inhibitors. The protein crystal structures were protonated, where hydrogen atoms were introduced with their 3D geometry. Furthermore, Swiss-PdbViewer was used to add missing amino acids and correct any faults discovered in the connection or type of distinct atoms.
Ligand Library preparation for molecular docking : Chemical structures of phytochemicals were obtained in the structure-data file (.sdf) format from the PubChem database (https://pub-chem.ncbi.nlm.nih.gov/). Using Open Babel, chemical structures of ligands in .sdf format were converted to .pdb format (O’Boyle et al., 2011). Ligand structures were generated by incorporating nonpolar hydrogens, Gasteiger modifications, and rotatable bonds, and then exported in .pdbqt format with the integration of AutoDock Vina 1.2.0 (Eberhardt et al., 2021).
Molecular Docking: Initially, the phytochemical structures were loaded with Alpha amylase and Acarbose target proteins using AutoDock Wizard. A three-dimensional grid box for target protein was generated using the MGL tools, and docking was accomplished with the PyRx-Python script V.0.8.(Oleg trott, 2012). Nine binding poses with both target proteins were produced for each ligand. The binding affinity was estimated as negative Gibbs free energy scores (kcal/mol), which used to rank the results.
The ADME (Absorption, Distribution, Metabolism, and Excretion) properties of the top-ranked phytochemical compounds were evaluated using the SwissADME web server (Daina, Michielin and Zoete, 2017) to assess drug-likeness parameters, including Lipinski’s rule of five, bioavailability score, and pharmacokinetic properties, while toxicity profiles were predicted using ProTox-II server (Banerjee et al., 2018) to determine LD50 values, toxicity classes, and potential adverse effects.
RESULTS AND DISCUSSION
A comprehensive phytochemical library comprising 3,320 compounds was successfully assembled from different sources, including 20 medicinal plants (Allium sativum, Aloe vera, Andrographis paniculata, Azadirachta indica, Cinnamomum verum, Cheilocostus speciosus, Costus speciosus, Eclipta prostrata, Ficus racemosa, Gymnema sylvestre, Mangifera indica, Momordica charantia, Ocimum sanctum, Ocimum tenuiflorum, Pterocarpus marsupium, Solanum americanum, Syzygium cumini, Trigonella foenum-graecum, and Zingiber officinale) and 24 synthetic anti-diabetic compounds (including Acetobexamide, Bezafibrate, Chlorpropanide, Dexamethasone, Duloxetine, Gilbenclamide, Gliciazide, Glimepiride, Glipizide, Glisoxepide, Glymidine, Hydrochlorothiazide, Irbesartan, Mazindol, Metformine, Mifepristone, Pentamidine, Phenformin, Pioglitazone, Prednisolone, Quinapril, Streptozocin, Tolazamide, Tolbutamide, and Troglitazone). The SMILES structures were retrieved from IMPPAT (1,412 compounds) and KNAPSACK (1,883 compounds) databases, with an additional 25 compounds obtained from PubChem standards. Acarbose was employed as a positive control for molecular docking validation against the α-amylase enzyme (PDB ID: 1B2Y).
Docking Study
Figure 1: Three-dimensional structure of protein 1b2y (center) surrounded by the chemical structures of ten ligand molecules: (1) Goyaglycoside-g, (2) Goyaglycoside-e, (3) Momordicoside R, (4) Kuguacin M, (5) beta-Amyrin, (6) alpha-Amyrin, (7) Melianoninol, (8) 13beta,28-Epoxy-11-ursen-3-one, (9) Odoratone, and (10) Acarbose. The figure illustrates the structural diversity of these ligands, which are investigated for their potential interactions with protein 1b2y in this study.
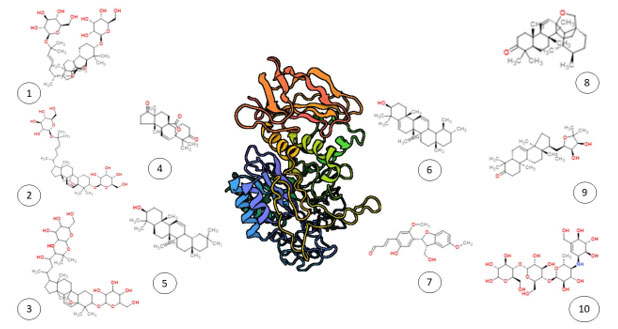
The molecular docking study revealed significant binding interactions between phytochemical compounds and the α-amylase enzyme 1B2Y (Figure 1). The comparative binding energy analysis (Figure 2) demonstrated that all ten top-ranked compounds exhibited superior binding affinities compared to the positive control Acarbose (-8.7 kcal/mol), with binding energies ranging from -11.7 to -18.6 kcal/mol.
Figure 2: Binding energies (kcal/mol) of the top ten molecules docked against alpha-amylase (protein 1b2y), shown alongside the positive control Acarbose.
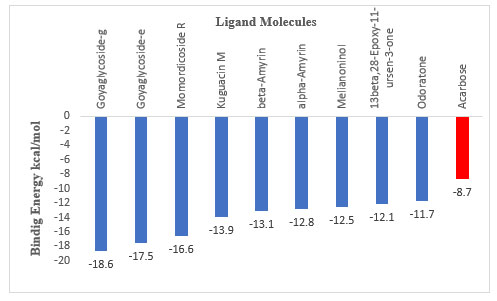
Goyaglycoside-g emerged as the most potent inhibitor with the highest binding affinity of -18.6 kcal/mol, representing a 2.14-fold improvement over Acarbose. Goyaglycoside-e ranked second with -17.5 kcal/mol (2.01-fold improvement), while Momordicoside R demonstrated strong binding at -16.6 kcal/mol (1.91-fold improvement).
The results demonstrate that natural compounds can significantly outperform the established inhibitor Acarbose, offering promising alternatives for diabetes management. The top-ranked compounds exhibited remarkable binding affinities ranging from -11.7 to -18.6 kcal/mol, substantially superior to Acarbose (-8.7 kcal/mol). Goyaglycoside-g showed the highest potency with a 2.14-fold improvement, while Momordicoside R demonstrated specific targeting of the catalytic residue TYR151, crucial for starch hydrolysis (Jayaraj, Suresh and Kadeppagari, 2013b). These findings align with previous studies emphasizing the effectiveness of plant-derived compounds in enzyme inhibition (Sales et al., 2012b; Kashtoh and Baek, 2022).
Table 1. Top ranked compounds from molecular docking analysis showing binding affinities, key molecular interactions, and significant residues involved in protein-ligand binding.
| Rank | Compound Name | Binding Affinity (kcal/mol) | Key Interactions | Significant Residues |
| 1 | Goyaglycoside-g | -18.6 | van der Waals | TRP58, TYR62 |
| 2 | Goyaglycoside-e | -17.5 | vdW + H-bond (LEU162) | LEU162 |
| 3 | Momordicoside R | -16.6 | vdW + TYR151 interaction | TYR151 |
| 4 | Kuguacin M | -13.9 | PI-Alkyl + vdW | TYR62, TRP58, ASP197, ASP300 |
| 5 | β-Amyrin | -13.1 | Pi-Sigma + Pi-Alkyl | Hydrophobic pocket |
| 6 | α-Amyrin | -12.8 | Alkyl | Hydrophobic pocket |
| 7 | Melianoninol | -12.5 | vdW + H-bond | A1.01, A2.24 |
| 8 | 13β,28-Epoxy-11-ursen-3-one | -12.1 | vdW | Hydrophobic pocket |
| 9 | Odoratone | -11.7 | vdW + H-bond + Pi-Alkyl | Hydrophobic/aromatic residues |
| 10 | Acarbose | -8.7 | vdW + PI-Alkyl | TRP58, TYR62, HIS305, LEU165 |
The detailed interaction analysis (Table 1) revealed distinct binding patterns for the moderately active compounds. Kuguacin M (-13.9 kcal/mol) exhibited excellent inhibitory potential through multiple PI-Alkyl interactions with TYR62, TRP58, ASP197, and ASP300, along with extensive van der Waals contacts. The triterpenoid β-Amyrin (-13.1 kcal/mol) demonstrated strong binding through Pi-Sigma and Pi-Alkyl interactions within the enzyme’s hydrophobic pocket, while α-Amyrin (-12.8 kcal/mol) showed similar alkyl-based interactions in the same hydrophobic region.
Figure 3: Binding poses and key residue contacts, highlighting van der Waals, Pi-Sigma, alkyl, and Pi-alkyl interactions for ligand Kuguacin M, beta-Amyrin, alpha-Amyrin, Melianoninol, Odoratone including the positive control Acarbose.
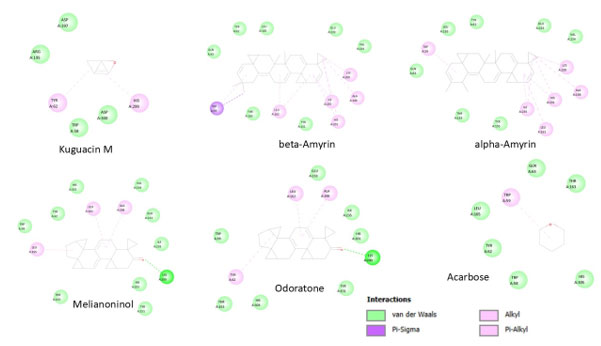
Melianoninol (-12.5 kcal/mol) displayed a balanced interaction profile combining van der Waals forces with hydrogen bonding capabilities, indicating stable enzyme-ligand complex formation. Odoratone (-11.7 kcal/mol) showed good inhibitory activity through a combination of van der Waals interactions, hydrogen bonds, and Pi-Alkyl contacts with both hydrophobic and aromatic residues.
Remarkably, these five compounds (Kuguacin M, β-Amyrin, α-Amyrin, Melianoninol, and Odoratone) demonstrated binding affinities 1.34 to 1.60-fold superior to Acarbose, indicating their excellent potential as natural α-amylase inhibitors (figure 3). The interaction profiles suggest that these compounds effectively occupy the enzyme’s active site through complementary molecular interactions, positioning them as promising anti-diabetic candidates with potentially fewer side effects than synthetic inhibitors.
The diverse interaction mechanisms observed, including van der Waals forces, hydrogen bonding, and Pi-Alkyl interactions, suggest stable enzyme-ligand complexes. The triterpenoids (β-Amyrin, α-Amyrin, and Kuguacin M) showed strong hydrophobic interactions, consistent with effective enzyme-ligand stability reported in previous studies (Salehi et al., 2019).
ADME Properties: The ADME (Absorption, Distribution, Metabolism, and Excretion) analysis of the phytochemical compounds revealed critical insights into their potential as anti-diabetic agents. Among the top candidates, Momordicoside R from Momordica charantia emerged as the most promising compound due to its strong binding affinity (-16.6 kcal/mol) targeting the catalytic residue TYR151 of α-amylase, which is crucial for inhibiting carbohydrate digestion. Despite its low gastrointestinal absorption and moderate solubility, Momordicoside R exhibits a favorable low logP (0.11), indicating better solubility compared to other glycosides like Goyaglycoside-g and -e. However, its high molecular weight and excessive hydrogen bond donors limit its bioavailability (score: 0.17), a common challenge for natural glycosides.
In contrast, Kuguacin M, another compound from Momordica charantia, shows high GI absorption and no Lipinski violations, making it more drug-like, but its hydrophobicity (logP: 3.63) and weaker binding affinity (-13.9 kcal/mol) reduce its therapeutic potential. Meanwhile, β-Amyrin and α-Amyrin, despite their strong hydrophobic interactions, suffer from poor solubility (logP >7) and low bioavailability, limiting their clinical applicability.
The control drug Acarbose, while highly soluble, has poor permeability and requires high doses, leading to gastrointestinal side effects. Momordicoside R’s targeted inhibition of TYR151 and natural origin make it a safer and more specific alternative. To overcome its bioavailability limitations, future research should focus on structural optimization (e.g., reducing molecular weight) and advanced formulations (e.g., nanoemulsions). In conclusion, Momordicoside R represents the best candidate for anti-diabetic development, provided its absorption challenges are addressed through further preclinical studies.
ADME analysis revealed that while Momordicoside R showed excellent binding affinity, its limited bioavailability (0.17) presents typical challenges for natural glycosides (Wang et al., 2015). However, its favorable logP value (0.11) indicates good solubility characteristics. Kuguacin M demonstrated better drug-like properties with high GI absorption and Lipinski compliance, despite moderate binding affinity.
Toxicity Prediction: The toxicity study of phytochemical compounds using ProTox-3.0 revealed important insights into their safety profiles as potential anti-diabetic agents. Among the evaluated compounds, Melianoninol emerged as a particularly promising candidate despite its moderate acute toxicity (LD50 = 2500 mg/kg, Class 5). While β-Amyrin and α-Amyrin showed the highest LD50 values (70,000 mg/kg, Class 6), indicating very low acute toxicity, their potential effects on GABA receptors and blood-brain barrier permeability raise concerns for therapeutic use. In contrast, Melianoninol demonstrated a more favorable overall toxicity profile, showing no hepatotoxicity or neurotoxicity, though it did present mild nephrotoxicity risk (0.61 probability) and significant immunotoxicity (0.98 probability). Importantly, Melianoninol’s toxicity profile compares favorably to the control drug Acarbose, which, despite having a higher LD50 (24,000 mg/kg), showed active hepatotoxicity, nephrotoxicity, and cardiotoxicity.
Melianoninol’s advantages include its natural origin, strong binding affinity to α-amylase (as shown in previous docking studies), and absence of severe organ toxicity risks. The main concerns for Melianoninol are its immunotoxicity potential and CYP450 inhibition (affecting CYP1A2, 2C19, 2C9, and 3A4), which could lead to drug-drug interactions in clinical use. However, these issues may be addressable through structural optimization and careful dosing strategies. When compared to other lead compounds, Melianoninol offers a balanced combination of efficacy and safety, making it a viable candidate for further development as a natural anti-diabetic agent.
Toxicity predictions identified Melianoninol as particularly promising, showing no hepatotoxicity or neurotoxicity compared to Acarbose, which exhibited multiple organ toxicity risks. This supports the general observation that natural compounds often display better safety profiles than synthetic drugs (Ardalani, Avan and Ghayour-Mobarhan, 2017; Omara et al., 2020).The identified phytochemicals address key limitations of current α-amylase inhibitors, including side effects and patient compliance issues (Kokil et al., 2010; El-Kaissi and Sherbeeni, 2011b). Compounds from Momordica charantia and Azadirachta indica validate traditional medicinal uses and support ethnopharmacological approaches to drug discovery.
CONCLUSION
The molecular docking studies presented in this research highlight the significant potential of phytochemical compounds as natural inhibitors of human α-amylase 1B2Y for managing type 2 diabetes mellitus. The findings demonstrate that plant-derived ligands exhibit superior binding affinities compared to the synthetic inhibitor Acarbose, with Goyaglycoside-g, Goyaglycoside-e, and Momordicoside R emerging as the most potent candidates. Notably, compounds derived from Momordica charantia (Kuguacin M, β-Amyrin, α-Amyrin) and Azadirachta indica (Melianoninol, Odoratone) showed remarkable inhibitory effects, underscoring the therapeutic promise of these plants. The interaction analysis revealed that these phytochemicals bind effectively to key residues of α-amylase, such as TYR151 and TRP58, through diverse mechanisms including van der Waals forces, hydrogen bonding, and hydrophobic interactions.
Despite challenges in bioavailability for some glycosides, their natural origin and favorable toxicity profiles position them as safer alternatives to synthetic drugs. This study validates the ethnopharmacological use of Momordica charantia and Azadirachta indica in traditional medicine and emphasizes their role as rich sources of anti-diabetic agents. Future research should focus on structural optimization and advanced formulations to enhance the bioavailability of these compounds. Overall, the findings advocate for the continued exploration of plant-based therapeutics in diabetes management, combining computational and experimental approaches to develop novel, effective, and safer treatments.
Conflict of Interest Statement: The authors declare that there are no conflicts of interest.
Author Contributions: Dhyey Kothari designed the study and wrote the manuscript. Manoj Godhaniya performed docking, data interpretation, and assisted with literature review. Kunjan Kikani supervised the work and revised the manuscript critically.
Funding Statement: No specific grant was received for this study.
Data Availability: All data are available from the corresponding author
Ethics Statement: This study does not involve human or animal participants; hence, ethical approval was not required.
REFERENCES
Omara, E. et al. (2010) ‘Herbal Medicines and Nutraceuticals for Diabetic Vascular Complications: Mechanisms of Action and Bioactive Phytochemicals’, Current Pharmaceutical Design, 16(34), pp. 3776–3807. Available at: https://doi.org/10.2174/138161210794455076.
Afendi, F.M. et al. (2012) ‘KNApSAcK family databases: integrated metabolite-plant species databases for multifaceted plant research’, Plant & Cell Physiology, 53(2), p. e1. Available at: https://doi.org/10.1093/pcp/pcr165.
Ardalani, H. et al. (2021) ‘Potential antidiabetic phytochemicals in plant roots: a review of in vivo studies’, Journal of Diabetes & Metabolic Disorders, 20(2), pp. 1837–1854. Available at: https://doi.org/10.1007/s40200-021-00853-9.
Ardalani, H., Avan, A. and Ghayour-Mobarhan, M. (2017) ‘Podophyllotoxin: a novel potential natural anticancer agent’, Avicenna Journal of Phytomedicine, 7(4), pp. 285–294.
Ayorinde, A.I. et al. (2025) ‘Molecular Docking, ADME and SAR Analysis of 383 Phytochemicals in the Quest for Lead Antidiabetic Inhibitors Targeting α-Amylase and α-Glucosidase Enzymes’, Tropical Journal of Drug Research, 2(1), p. 6. Available at: https://doi.org/10.26538/tjdr/v2i1.2.
Banerjee, P. et al. (2018) ‘ProTox-II: a webserver for the prediction of toxicity of chemicals’, Nucleic Acids Research, 46(W1), pp. W257–W263. Available at: https://doi.org/10.1093/nar/gky318.
Bentham Science Publisher, B.S.P. (2006) ‘Methods for the Prediction of Protein-Ligand Binding Sites for Structure-Based Drug Design and Virtual Ligand Screening.’, Current Protein & Peptide Science, 7(5), pp. 395–406. Available at: https://doi.org/10.2174/138920306778559386.
Daina, A., Michielin, O. and Zoete, V. (2017) ‘SwissADME: a free web tool to evaluate pharmacokinetics, drug-likeness and medicinal chemistry friendliness of small molecules’, Scientific Reports, 7(1), p. 42717. Available at: https://doi.org/10.1038/srep42717.
DeFronzo, R.A. et al. (2015) ‘Type 2 diabetes mellitus’, Nature Reviews Disease Primers, 1(1), p. 15019. Available at: https://doi.org/10.1038/nrdp.2015.19.
Devakrishna, K., Taj, G. and Upadhyay, S. (2024) ‘In silico molecular docking analysis of Cichorium intybus L. phytochemical compounds against two related targets of type 2 diabetes mellitus’, Annals of Phytomedicine An International Journal, 13(1). Available at: https://doi.org/10.54085/ap.2024.13.1.60.
Eberhardt, J. et al. (2021) ‘AutoDock Vina 1.2.0: New Docking Methods, Expanded Force Field, and Python Bindings’, Journal of Chemical Information and Modeling, 61(8), pp. 3891–3898. Available at: https://doi.org/10.1021/acs.jcim.1c00203.
El-Kaissi, S. and Sherbeeni, S. (2011a) ‘Pharmacological Management of Type 2 Diabetes Mellitus: An Update’, Current Diabetes Reviews, 7(6), pp. 392–405. Available at: https://doi.org/10.2174/157339911797579160.
El-Kaissi, S. and Sherbeeni, S. (2011b) ‘Pharmacological management of type 2 diabetes mellitus: an update’, Current Diabetes Reviews, 7(6), pp. 392–405. Available at: https://doi.org/10.2174/157339911797579160.
Grubelnik, V. et al. (2020) ‘Mitochondrial Dysfunction in Pancreatic Alpha and Beta Cells Associated with Type 2 Diabetes Mellitus’, Life, 10(12), p. 348. Available at: https://doi.org/10.3390/life10120348.
Jayaraj, S., Suresh, S. and Kadeppagari, R. (2013a) ‘Amylase inhibitors and their biomedical applications’, Starch – Stärke, 65(7–8), pp. 535–542. Available at: https://doi.org/10.1002/star.201200194.
Jayaraj, S., Suresh, S. and Kadeppagari, R. (2013b) ‘Amylase inhibitors and their biomedical applications’, Starch – Stärke, 65(7–8), pp. 535–542. Available at: https://doi.org/10.1002/star.201200194.
Kanter, J.E. and Bornfeldt, K.E. (2016) ‘Impact of Diabetes Mellitus’, Arteriosclerosis, Thrombosis, and Vascular Biology, 36(6), pp. 1049–1053. Available at: https://doi.org/10.1161/ATVBAHA.116.307302.
Kashtoh, H. and Baek, K.-H. (2022) ‘Recent Updates on Phytoconstituent Alpha-Glucosidase Inhibitors: An Approach towards the Treatment of Type Two Diabetes’, Plants, 11(20), p. 2722. Available at: https://doi.org/10.3390/plants11202722.
Kashtoh, H. and Baek, K.-H. (2023) ‘New Insights into the Latest Advancement in α-Amylase Inhibitors of Plant Origin with Anti-Diabetic Effects’, Plants, 12(16), p. 2944. Available at: https://doi.org/10.3390/plants12162944.
Kaur, N. et al. (2021) ‘Alpha‐amylase as molecular target for treatment of diabetes mellitus: A comprehensive review’, Chemical Biology & Drug Design, 98(4), pp. 539–560. Available at: https://doi.org/10.1111/cbdd.13909.
Khan, M.A.B. et al. (2019) ‘Epidemiology of Type 2 Diabetes – Global Burden of Disease and Forecasted Trends’:, Journal of Epidemiology and Global Health, 10(1), p. 107. Available at: https://doi.org/10.2991/jegh.k.191028.001.
Kim, S. et al. (2025) ‘PubChem 2025 update’, Nucleic Acids Research, 53(D1), pp. D1516–D1525. Available at: https://doi.org/10.1093/nar/gkae1059.
Kokil, G.R. et al. (2010) ‘Pharmacology and chemistry of diabetes mellitus and antidiabetic drugs: a critical review’, Current Medicinal Chemistry, 17(35), pp. 4405–4423. Available at: https://doi.org/10.2174/092986710793361225.
Kokil, G.R. et al. (2015) ‘Type 2 Diabetes Mellitus: Limitations of Conventional Therapies and Intervention with Nucleic Acid-Based Therapeutics’, Chemical Reviews, 115(11), pp. 4719–4743. Available at: https://doi.org/10.1021/cr5002832.
Kurniawan, K., Marfu’ah, N. and Fazriah D., G. (2025) ‘In Silico Study of Gamma-Mangostin Compound from Garcinia Mangostana L. Fruit Skin and Activity Test as an Alpha Amylase Inhibitor’, Journal La Medihealtico, 6(2), pp. 395–410. Available at: https://doi.org/10.37899/journallamedihealtico.v6i2.1889.
Li, H. et al. (2021) ‘Proteinaceous α-amylase inhibitors: purification, detection methods, types and mechanisms’, International Journal of Food Properties, 24(1), pp. 277–290. Available at: https://doi.org/10.1080/10942912.2021.1876087.
Mathur, N. et al. (2024) ‘In Silico Docking: Protocols for Computational Exploration of Molecular Interactions’, in Č. Podlipnik (ed.) Biomedical Engineering. IntechOpen. Available at: https://doi.org/10.5772/intechopen.1005527.
McMillan, J., Bester, M.J. and Apostolides, Z. (2025) ‘In silico docking and ADMET studies on clinical targets for type 2 diabetes correlated to in vitro inhibition of pancreatic alpha-amylase and alpha-glucosidase by rutin, caffeic acid, p-coumaric acid, and vanillin’, In Silico Pharmacology, 13(1), p. 42. Available at: https://doi.org/10.1007/s40203-025-00324-6.
Mikłosz, A. and Chabowski, A. (2023) ‘Adipose-derived Mesenchymal Stem Cells Therapy as a new Treatment Option for Diabetes Mellitus’, The Journal of Clinical Endocrinology & Metabolism, 108(8), pp. 1889–1897. Available at: https://doi.org/10.1210/clinem/dgad142.
Mohanraj, K. et al. (2018) ‘IMPPAT: A curated database of Indian Medicinal Plants, Phytochemistry And Therapeutics’, Scientific Reports, 8(1), p. 4329. Available at: https://doi.org/10.1038/s41598-018-22631-z.
Mursal, M. et al. (2024a) ‘Navigating the Computational Seas: A Comprehensive Overview of Molecular Docking Software in Drug Discovery’, in Č. Podlipnik (ed.) Biomedical Engineering. IntechOpen. Available at: https://doi.org/10.5772/intechopen.1004802.
Mursal, M. et al. (2024b) ‘Navigating the Computational Seas: A Comprehensive Overview of Molecular Docking Software in Drug Discovery’, in Č. Podlipnik (ed.) Biomedical Engineering. IntechOpen. Available at: https://doi.org/10.5772/intechopen.1004802.
O’Boyle, N.M. et al. (2011) ‘Open Babel: An open chemical toolbox – 1758-2946-3-33.pdf’, Journal of Cheminformatics, 3(33), pp. 1–14.
Oleg trott, A.J.O. (2012) ‘AutoDock Vina: Improving the Speed and Accuracy of Docking with a New Scoring Function, Efficient Optimization, and Multithreading’, Journal of computational chemistry, 32, pp. 174–182. Available at: https://doi.org/10.1002/jcc.
Omara, T. et al. (2020) ‘Medicinal Plants Used in Traditional Management of Cancer in Uganda: A Review of Ethnobotanical Surveys, Phytochemistry, and Anticancer Studies’, Evidence-Based Complementary and Alternative Medicine: eCAM, 2020, p. 3529081. Available at: https://doi.org/10.1155/2020/3529081.
Patel, J.R. et al. (2022) ‘A Review on Computational Software Tools for Drug Design and Discovery’, Indo Global Journal of Pharmaceutical Sciences, 12, pp. 53–81. Available at: https://doi.org/10.35652/IGJPS.2022.12006.
Pinzi, L. and Rastelli, G. (2019) ‘Molecular Docking: Shifting Paradigms in Drug Discovery’, International Journal of Molecular Sciences, 20(18), p. 4331. Available at: https://doi.org/10.3390/ijms20184331.
Salehi, B. et al. (2019) ‘Antidiabetic Potential of Medicinal Plants and Their Active Components’, Biomolecules, 9(10), p. 551. Available at: https://doi.org/10.3390/biom9100551.
Sales, P.M. et al. (2012a) ‘α-Amylase Inhibitors: A Review of Raw Material and Isolated Compounds from Plant Source’, Journal of Pharmacy & Pharmaceutical Sciences, 15(1), p. 141. Available at: https://doi.org/10.18433/J35S3K.
Sales, P.M. et al. (2012b) ‘α-Amylase Inhibitors: A Review of Raw Material and Isolated Compounds from Plant Source’, Journal of Pharmacy & Pharmaceutical Sciences, 15(1), p. 141. Available at: https://doi.org/10.18433/J35S3K.
Schneuing, A. et al. (2024) ‘Structure-based drug design with equivariant diffusion models’, Nature Computational Science, 4(12), pp. 899–909. Available at: https://doi.org/10.1038/s43588-024-00737-x.
Silva, B. et al. (2017a) ‘Promising Potential of Dietary (Poly)Phenolic Compounds in the Prevention and Treatment of Diabetes Mellitus’, Current Medicinal Chemistry, 24(4), pp. 334–354. Available at: https://doi.org/10.2174/0929867323666160905150419.
Silva, B. et al. (2017b) ‘Promising Potential of Dietary (Poly)Phenolic Compounds in the Prevention and Treatment of Diabetes Mellitus’, Current Medicinal Chemistry, 24(4), pp. 334–354. Available at: https://doi.org/10.2174/0929867323666160905150419.
Varshney, M. et al. (2024) ‘In Silico and In Vitro Alpha-amylase Activities of Previously Synthesized Pyridazine Derivatives’, Philippine Journal of Science, 153(3). Available at: https://doi.org/10.56899/153.03.31.
Vinayagam, R., Xiao, J. and Xu, B. (2017a) ‘An insight into anti-diabetic properties of dietary phytochemicals’, Phytochemistry Reviews, 16(3), pp. 535–553. Available at: https://doi.org/10.1007/s11101-017-9496-2.
Vinayagam, R., Xiao, J. and Xu, B. (2017b) ‘An insight into anti-diabetic properties of dietary phytochemicals’, Phytochemistry Reviews, 16(3), pp. 535–553. Available at: https://doi.org/10.1007/s11101-017-9496-2.
Vollenweider, P., Von Eckardstein, A. and Widmann, C. (2015) ‘HDLs, Diabetes, and Metabolic Syndrome’, in A. Von Eckardstein and D. Kardassis (eds) High Density Lipoproteins. Cham: Springer International Publishing (Handbook of Experimental Pharmacology), pp. 405–421. Available at: https://doi.org/10.1007/978-3-319-09665-0_12.
Wang, Y. et al. (2015) ‘In silico ADME/T modelling for rational drug design’, Quarterly Reviews of Biophysics, 48(4), pp. 488–515. Available at: https://doi.org/10.1017/S0033583515000190.


