Department of Biological Science, Faculty of Science, King Abdulaziz University, Jeddah, Kingdom of Saudi Arabia
Corresponding author email: ahmadfirozbin@gmail.com
Article Publishing History
Received: 27/11/2020
Accepted After Revision: 21/03/2021
The proprotein convertases (PCs) are involved in variety of cellular precursors in the secretory pathway. Polymorphisms in proprotein convertase subtilisin/kexin type 1 (PCSK1) have been associated with adult and childhood obesity. In this work non synonymous SNPs of the PCSK1 gene were retrieved from the dbSNP database. In order to predict the damaging or deleterious nsSNPs, multiple consensus tools were employed by using online tool VEP. Further we also employed SNP-GO tools to predict pathogenic nonsynonymous SNPs. Mutants like D176Y, E345A, G228V, G308E, G310R, G440E, G442R, R110C, S382L, W130S and W404R have shown deleterious and highest pathogenicity. These predicted deleterious and pathogenic nsSNPs are expected to have impending functional influence and may contribute in understanding the functional roles of PCSK1 gene associated with obesity.
nsSNP, Proprotein convertase subtilisin/kexin type 1, Neuroendocrine convertase 1, In Silico Analysis, PCSK1
Majeed. A. H. M, Ali H. M , Anwar. Y, Ullah. I, Firoz. A. In-Silico Analysis of Nonsynonymous Single Nucleotide Polymorphism in Human PCSK1 gene. Biosc.Biotech.Res.Comm. 2021;14(1).
Majeed. A. H. M, Ali H. M , Anwar. Y, Ullah. I, Firoz. A. In-Silico Analysis of Nonsynonymous Single Nucleotide Polymorphism in Human PCSK1 gene. Biosc.Biotech.Res.Comm. 2021;14(1). Available from: <a href=”https://bit.ly/3s2zAiL“>https://bit.ly/3s2zAiL</a>
Copyright © Majeed et al., This is an open access article distributed under the terms of the Creative Commons Attribution License (CC-BY) https://creativecommns.org/licenses/by/4.0/, which permits unrestricted use distribution and reproduction in any medium, provide the original author and source are credited.
INTRODUCTION
An obesity increasing worldwide and polymorphisms in proprotein convertase subtilisin/kexin type 1 (PCSK1) gene have been associated with adult and childhood obesity. Body mass index variation (risk of common obesity) is associated with more than 60 single-nucleotide polymorphisms (SNPs), identified by genome-wide association studies (Philippe 2015). The proprotein convertases (PCs) are involved in variety of cellular precursors in the secretory pathway and due to homology of their catalytic domains to bacterial subtilisin and yeast kexin, the genes are known as subtilisin and kexin-like proprotein convertases (PCSKs) (Stijnen, 2016 Loffler , 2016). Human PCSK1 gene consists of 14 exons located on chromosome 5 (Ramos-Molina, 2016), and its promoter contains transcriptional elements CRE-1 and CRE-2 which can be transactivated by CREB-1 and ATF1 transcription factors (Espinosa, 2008; Stijnen,2016).
Analysis of human tissues and cells revealed the presence of a dominant transcript and the major sites of expression being endocrine pancreas, pituitary and brain (Stijnen, 2016). 71% of PCSK1 variant were located in coding region of the catalytic domain and 21% are located on the P domain (Akinci 2019). Many studies show a strong evidence about rs6232 and rs6235 involving with obesity (Jackson ,2003). association with three variants are found in PCSK1 gene rs6232 encoding by N221D substitution involve in reduce the activity of PC1/3 while rs234 encodes by Q665E compatible with rs6235 that encodes by S690T are essential to form a linkage between PC1/3 and its sorting in secretory granules (Stijnen, 2016; Frank 2013),
These variants have a significantly role in reducing the level of plasma glucose rapidly and increase serum insulin levels causing a hyperglycemia as type 2 diabetes (T2D) by increasing in glocuse production, insulin resistance and a dysfunction in β cell that found in pancreatic cells (Gjesing, 2011), along with the effect of blood pressure and energy ratio, causing a hypertension in the blood vessels which lead to a cardiovascular (Heni, 2010 Pepin et al 2019).The R405X mutation cause a deletion of P and C-terminal tail domain (Bandsma, 2013).
Identified N309K a deleterious in PCSK1 gene which make C-terminal domain incapable of cleave in intermolecular interaction (Wilschanski, 2014). K26E is located before the signal peptide cleavage site, M125I, T175M, N180S, Y181H, G226R and S325N are located in the catalytic domain and the T558A is located in the middle domain. These mutations have an impact on PC1/3 folding and its stability. also, G209R and G593R mutation might affect on the PC1/3 misfolding due to their enzymatic activation (Blanco, 2015).In addition, T175M was defined as induce the inhibition in N-glycosylation site which is responsible for cellular signal and altering the protein maturation (Creemers, 2012 Pepin et al 2019).
MATERIAL AND METHODS
Datasets:The SNPs of the PCSK1 gene were retrieved from the dbSNP database (Sherry, 2001). Keyword “Human PCSK1” used as our search term. Furthermore, it is filtered by selecting variation class as SNV, function class as missense. The protein sequences (P29120) were retrieved from the UniProt (https://www.uniprot.org)
Predicting deleterious and damaging nsSNPs: In order to predict the damaging or deleterious nsSNPs, multiple consensus tools were employed by using online tool VEP (http://www.ensembl.org/Tools/VEP). VEP advantages include: it uses latest human genome assembly GRCh38.p10, and can predict thousands of SNPs from multiple tools including SIFT, PROVEAN, Condel, and PolyPhen-2, at a time. nsSNP rs-ids were uploaded to VEP tool to get the prediction results
SIFT: The algorithm predicted that the tolerant and intolerant coding base substitution based upon properties of amino acids and homology of sequence (Choi Y, 2015). The tool considered that vital positions in the protein sequence have been conserved throughout evolution and therefore substitutions at conserved alignment position is expected to be less tolerated and affect protein function than those at diverse positions., SIFT predicted substituted amino acid as damaging at default threshold score <0.05, while score ³ 0.05 is predicted as tolerated.
PolyPhen-2: This tool is predicting the structural and functional consequences of a particular amino acid substitution in human protein (Adzhubei, 2010). Prediction of PolyPhen-2 is based on a number of features including information of structural and sequence comparison. The PolyPhen-2 score varies between 0.0 (benign) to 10.0 (damaging). The PolyPhen-2 prediction output categorizes the SNPs into three basic categories, benign (score < 0.2), possibly damaging, (score between 0.2 and0.96), or probably damaging (score >0.96).
Provean: This tool (http://provean.jcvi.org/) uses an alignment-based scoring method for predicting the functional consequences of single and multiple amino acid substitutions, and in-frame deletions and insertions (Choi, 2015). The tool has a default threshold score, i.e. -2.5, below which a protein variant is predicted as deleterious, and above that threshold, a protein variant is neutral.
CONDEL (CONsensus DELeteriousness): This tool evaluates the probability of missense single nucleotide variants (SNVs) deleterious. it computes a weighted average of the scores of SIFT, PolyPhen2, MutationAssessor and FatHMM (Hecht , 2015).
Predicting disease associated nsSNPs
SNPs &GO: A web server predicting whether an amino acid substitution is associated to a disease or not (http://snps.biofold.org/snps-and-go) (). It is a SVM (Support Vector Machine) based tool which takes features of protein sequence, evolutionary information, and functional annotation according to Gene Ontology terms. We input isoform 1 of Swiss-Prot Code of LSP1 (P33241) and provided the list of amino acid mutations. The results predicted the probability for the polymorphisms of helicase whether being disease- associated or not by three methods: (a) SNPs&GO, (b) PhD-SNP, and (c) PANTHER. Probability score >0.5 is predicted as disease associated variation (Calabrese , 2015).
RESULTS AND DISCUSSION
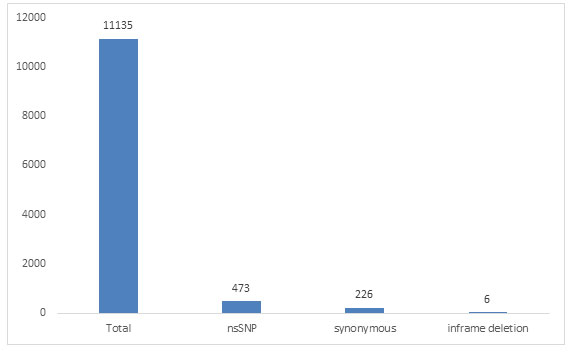
473 nsSNP ids of human PCSK1 gene was downloaded from dbSNP database of NCBI (Supplementary Table 1), after filtering variation class SNV, function class missense, there were 473 SNP mapped to missense, 226 SNPs mapped to synonymous and 6 SNPs mapped to inframe deletion, while 11135 mapped to total SNPs of different variation class (Figure 1).
Figure 1: Number of SNPs in different function class of PCSK1 gene of human from dbSNP database
Predicting deleterious and damaging pathogenic nsSNPs: In order to predict the damaging or deleterious nsSNPs multiple consensus tools were employed. Initially, online tool VEP was used (McLaren, 2016). VEP advantages include: it uses latest human genome assembly GRCh38.p10, and can predict thousands of SNPs from multiple tools including SIFT, Condel, and PolyPhen-2, at a time. 473 nsSNP rsids were uploaded to VEP tool and the prediction results were taken on default scores of consensus tools based on sequence and structure homology methods: (a) SIFT (score <-0.5) (b) Polyphen (score >0.96) (c) PROVEAN (score< 2.5) and Condel (score >0.522).
In order to get a very high confident nsSNPs impacting structure and function of PCSK1 gene, 46 nsSNPs out of 473 nsSNP (Table 1) were found to be deleterious by all four tools and predicted disease by panther tools, and these eleven nsSNPs rs552958813 of mutation D176Y, rs864309557, of mutation E345A, rs747169606 of mutation G228V, rs990328651 of mutation G308E, rs748808191 of mutation G310R, rs865777271 of mutation G440E, rs761336991 of mutation G442R, rs774036542 of mutation R110C, rs1561368007 of mutation S382L, rs1434467255 of mutation W130S and rs1180593976 of mutation W404R were predicted highly pathogenic with more than 9 RI score (Table-1).
Table 1. 46 Predicted deleterious and pathogenic missense SNPs of PCSK1 gene using prediction tools such as SIFT, Condel, Polyphen and PROVEAN and PANTHER.
| SNP-ids | AA-change | SIFT (score) | PolyPhen (score) | Condel score) | PROVEAN (score) | PANTHER Prediction | RI |
| rs759379849 | D193G | *(0) | #(0.999) | *(0.935) | * (0.92173) | Disease | 8 |
| rs1561374455 | D195G | *(0) | #(0.96) | *(0.848) | *(0.91956) | Disease | 8 |
| rs752416942 | D272G | *(0) | #(1) | *(0.945) | *(0.93175) | Disease | 8 |
| rs749888385 | T353I | *(0.02) | #(0.967) | *(0.792) | *(0.78636) | Disease | 8 |
| rs762403860 | A213V | *(0) | #(0.998) | *(0.919) | *(0.71639) | Disease | 9 |
| rs1475050973 | C212R | *(0) | #(1) | *(0.945) | *(0.99425) | Disease | 9 |
| rs552958813 | D176N | *(0) | #(1) | *(0.945) | *(0.80172) | Disease | 9 |
| rs752416942 | D272V | *(0) | #(1) | *(0.945) | *(0.9783) | Disease | 9 |
| rs1363728113 | G155D | *(0) | #(0.98) | *(0.869) | *(0.90023) | Disease | 9 |
| rs1382566997 | G155S | *(0) | #(0.986) | *(0.879) | *(0.83899) | Disease | 9 |
| rs1490377137 | G158A | *(0) | #(0.999) | *(0.935) | *(0.873) | Disease | 9 |
| rs768031892 | G209R | *(0) | #(1) | *(0.945) | *(0.95336) | Disease | 9 |
| rs142673134 | G279A | *(0.04) | #(0.959) | *(0.752) | *(0.86296) | Disease | 9 |
| rs1312543959 | G298A | *(0) | #(0.999) | *(0.935) | *(0.87223) | Disease | 9 |
| rs778681269 | G311R | *(0) | #(1) | *(0.945) | *(0.95276) | Disease | 9 |
| rs567641208 | G390S | *(0) | #(0.999) | *(0.935) | *(0.88839) | Disease | 9 |
| rs1389330621 | N180K | *(0) | #(0.999) | *(0.935) | *(0.85994) | Disease | 9 |
| rs1269967613 | N429K | *(0) | #(0.994) | *(0.897) | *(0.87835) | Disease | 9 |
| rs1246203022 | P280S | *(0) | #(1) | *(0.945) | *(0.96058) | Disease | 9 |
| rs775618000 | P341L | *(0) | #(1) | *(0.945) | *(0.98437) | Disease | 9 |
| rs775136858 | P386L | *(0) | #(0.998) | *(0.919) | *(0.98692) | Disease | 9 |
| rs1332430207 | Q408R | *(0) | #(1) | *(0.945) | *(0.73267) | Disease | 9 |
| rs748072514 | R110H | *(0) | #(0.999) | *(0.935) | *(0.76822) | Disease | 9 |
| rs768934109 | R296I | *(0) | #(1) | *(0.945) | *(0.95246) | Disease | 9 |
| rs1421014042 | S186N | *(0) | #(0.996) | *(0.906) | *(0.59873) | Disease | 9 |
| rs137852824 | S307L | *(0) | #(0.999) | *(0.935) | *(0.86222) | Disease | 9 |
| rs1166018774 | T210S | *(0) | #(0.999) | *(0.935) | *(0.71762) | Disease | 9 |
| rs1303515025 | T276I | *(0) | #(0.996) | *(0.906) | *(0.84742) | Disease | 9 |
| rs766414747 | T375K | *(0) | #(0.998) | *(0.919) | *(0.88839) | Disease | 9 |
| rs766414747 | T375M | *(0) | #(0.993) | *(0.895) | *(0.88839) | Disease | 9 |
| rs1346360455 | T381I | *(0) | #(1) | *(0.945) | *(0.88839) | Disease | 9 |
| rs1434467255 | W130L | *(0) | #(1) | *(0.945) | *(0.99433) | Disease | 9 |
| rs868424536 | W152L | *(0) | #(0.985) | *(0.877) | *(0.99023) | Disease | 9 |
| rs1245583638 | W342G | *(0) | #(0.998) | *(0.919) | *(0.99704) | Disease | 9 |
| rs1246742230 | W98R | *(0.02) | #(0.994) | *(0.835) | *(0.99587) | Disease | 9 |
| rs552958813 | D176Y | *(0) | #(1) | *(0.945) | *(0.97364) | Disease | 10 |
| rs864309557 | E345A | *(0) | #(1) | *(0.945) | *(0.873) | Disease | 10 |
| rs747169606 | G228V | *(0) | #(1) | *(0.945) | *(0.97617) | Disease | 10 |
| rs990328651 | G308E | *(0) | #(1) | *(0.945) | *(0.95246) | Disease | 10 |
| rs748808191 | G310R | *(0) | #(1) | *(0.945) | *(0.95276) | Disease | 10 |
| rs865777271 | G440E | *(0) | #(1) | *(0.945) | *(0.95665) | Disease | 10 |
| rs761336991 | G442R | *(0) | #(0.999) | *(0.935) | *(0.95665) | Disease | 10 |
| rs774036542 | R110C | *(0) | #(1) | *(0.945) | *(0.93582) | Disease | 10 |
| rs1561368007 | S382L | *(0) | #(0.998) | *(0.919) | *(0.88839) | Disease | 10 |
| rs1434467255 | W130S | *(0) | #(1) | *(0.945) | *(0.99699) | Disease | 10 |
| rs1180593976 | W404R | *(0) | #(0.998) | *(0.919) | *(0.99969) | Disease | 10
|
(*Deleterious, #Probably Damaging)
Studies show a strong evidence about variants are found in PCSK1 gene involving with obesity, association with variants N221D, S690T and Q665E substitutions found in PCSK1 gene involve in reduce the activity of PC1/3, linkage between PC1/3 and its sorting in secretory granules ( Jackson 2003, Stijnen 2016; Frank 2013), Identified N309K a deleterious in PCSK1 gene which make C-terminal domain incapable of cleave in intermolecular interaction (Wilschanski, 2014). K26E is located before the signal peptide cleavage site, M125I, T175M, N180S, Y181H, G226R and S325N are located in the catalytic domain and the T558A is located in the middle domain.
These mutations have an impact on PC1/3 folding and its stability. also, G209R and G593R mutation might affect on the PC1/3 misfolding due to their enzymatic activation (Blanco EH, 2015). In addition, T175M was defined as induce the inhibition in N-glycosylation site which is responsible for cellular signal and altering the protein maturation (Creemers, 2012). Pickett had proposed that R80Q have the most influence part in PC1/3 maturation and its activity (Pickett, 2013). In another report, the S357G mutant that low the calcium dependence and highly resistance the peptide inhibitors (Blanco, 2015).
CONCLUSION
Our investigation shows mutants D176Y, E345A, G228V, G308E, G310R, G440E, G442R, R110C, S382L, W130S and W404R with deleterious and highest pathogenicity, and may offer valuable information in selecting SNPs that are expected to have impending functional influence and pathogenicity also eventually may contribute in understanding the functional roles of PCSK1 gene associated with obesity.
ACKNOWLEDGMENTS
This work was not supported by any funding agency. We acknowledge with thanks Bioinformatics and Computational Biology Unit at Department of Biological Sciences King Abdulaziz University, Jeddah, KSA for providing their support and facilities.
REFERENCES
Adzhubei IA, Schmidt S, Peshkin L, et al (2010) A method and server for predicting damaging missense mutations.Nat Methods.7(4):248-9.
Akıncı A, Türkkahraman D, Tekedereli İ et al (2019) Novel Mutations in Obesity-related Genes in Turkish Children with Non-syndromic Early Onset Severe Obesity: A Multicentre Study. J Clin Res Pediatr Endocrinol. 22;11(4):341-349.
Bandsma RH, Sokollik C, Chami R, et al (2013). From diarrhoea to obesity in prohormone convertase 1/3 deficiency: age-dependent clinical, pathologic, and enteroendocrine characteristics. J Clin Gastroenterol. 47(10):834-843.
Blanco EH, Ramos-Molina B, Lindberg I (2015). Revisiting PC1/3 Mutants: Dominant-Negative Effect of Endoplasmic Reticulum-Retained Mutants. Endocrinology. 156(10):3625-3637.
Calabrese R, Capriotti E, Fariselli P, Martelli PL, Casadio R (2009). Functional annotations improve the predictive score of human disease-related mutations in proteins. Hum Mutat. 30(8):1237-44.
Choi Y, Chan AP (2015). PROVEAN web server: a tool to predict the functional effect of amino acid substitutions and indels. Bioinformatics.15;31(16):2745-7
Creemers JW, Choquet H, Stijnen P et al (2012); Heterozygous mutations causing partial prohormone convertase 1 deficiency contribute to human obesity. Diabetes. 61(2):383-90
El-Sayed Moustafa JS, Froguel P et al (2013). From obesity genetics to the future of personalized obesity therapy. Nat Rev Endocrinol 9: 402–413
Espinosa VP, Liu Y, Ferrini M, et al (2008). Differential regulation of 37 prohormone convertase 1/3, prohormone convertase 2 and phosphorylated cyclic38 AMP-response element binding protein by short-term and long-term morphine 39 treatment: implications for understanding the “switch” to opiate addiction. 40 Neuroscience 156(3):788–99.
Frank GR, Fox J, Candela N, et al (2013). Severe obesity and diabetes insipidus in a patient with PCSK1 deficiency. Mol Genet Metab. 110(1-2):191-194.
Gjesing AP, Vestmar MA, Jørgensen Tet al (2011). The effect of PCSK1 variants on waist, waist-hip ratio and glucose metabolism is modified by sex and glucose tolerance status. PLoS One.6(9):e23907
Harter B, Fuchs I, Müller T, Akbulut UE, Cakir M, Janecke AR (2016). Early Clinical Diagnosis of PC1/3 Deficiency in a Patient With a Novel Homozygous PCSK1 Splice-Site Mutation. J Pediatr Gastroenterol Nutr.62(4):577-80.
Hecht M., Bromberg, Rost, B (2015). Better prediction of functional effects for sequence variants. BMC Genomics 16, S1.
Heni M, Haupt A, Schäfer SA. et al (2010). Association of obesity risk SNPs in PCSK1with insulin sensitivity and proinsulin conversion. BMC Med Genet 11, 86 (2010).
Jackson RS, Creemers JW, Farooqi IS, et al; Small-intestinal dysfunction accompanies the complex endocrinopathy of human proprotein convertase 1 deficiency. J Clin Invest. 2003 Nov;112(10):1550-60.
McLaren W, Gil L, Hunt SE, Riat HS, Ritchie GR, Thormann A, Flicek P, Cunningham F (2016) The Ensembl Variant Effect Predictor. Genome Biology Jun 6;17(1):122.
Pepin L, Colin E, Tessarech M et al; (2019) A New Case of PCSK1 Pathogenic Variant With Congenital Proprotein Convertase 1/3 Deficiency and Literature Review. J Clin Endocrinol Metab. Apr 1;104(4):985-993.
Philippe, J., Stijnen, P., Meyre, D. et al. (2015) A nonsense loss-of-function mutation in PCSK1 contributes to dominantly inherited human obesity. Int J Obes 39, 295–302. https://doi.org/10.1038/ijo.2014.96
Pickett LA, Yourshaw M, Albornoz V, Chen Z, Solorzano-Vargas RS, Nelson SF, et al. (2013) Functional Consequences of a Novel Variant of PCSK1. PLoS ONE 8(1): e55065.
Ramos-Molina B, Martin MG, Lindberg I.(2016) PCSK1 Variants and Human Obesity. Prog Mol Biol Transl Sci. 140:47-74. doi: 10.1016/bs.pmbts.2015.12.001. Epub 2016 Jan 29.
Sherry ST, Ward MH, Kholodov M, et al.(2001) dbSNP: the NCBI database of genetic variation. Nucleic Acids Res. 29(1):308-311.
Stijnen P, Ramos-Molina B, O’Rahilly S, Creemers John W. M.,(2016) PCSK1 Mutations and Human Endocrinopathies: From Obesity to Gastrointestinal Disorders, Endocrine Reviews, Volume 37, Issue 4, Pages 347–371.
Wilschanski M, Abbasi M, Blanco E, Lindberg I, Yourshaw M, Zangen D, et al. (2014) A Novel Familial Mutation in the PCSK1 Gene That Alters the Oxyanion Hole Residue of Proprotein Convertase 1/3 and Impairs Its Enzymatic Activity. PLoS ONE 9(10).


