1Department of Biotechnology, Faculty of Life Sciences and Technology, AKS University, Satna, Madhya Pradesh, India
2Forest Genetics, Plant Propagation and Biotechnology State Forest Research Institute, Jabalpur Madhya Pradesh, India
Corresponding author email: biotech.yadav0@gmail.com
Article Publishing History
Received: 05/11/2020
Accepted After Revision: 19/12/2020
Saraca asoca (Roxb.) De Wilde is considered as a medicinally important plant species with higher therapeutic value. It has immense therapeutic value due to presence of various bioactive compounds in it. Geographical locations play important roles in the production of pharmaceutically important compounds and this variation comes from the genetic background of the plants. S. asoca is found at different locations of India and may differ in their genetic architecture. Due to high pharmaceutical values it is important to explore diversity among genotypes available in India. The present investigation was carried out to explore inherent diversity at DNA level among 15 S. asoca genotypes collected from different geographical locations of India. For this purpose a total of 40 inter simple sequence repeat markers (ISSR) were used. Out of 40 ISSR markers, only 28 markers produced polymorphic bands.
The total numbers of bands amplified were 149 out of them 133 bands were found to be polymorphic. A dendrogram was generated on the basis of UPGMA clustering. All S. asoca genotypes were clustered into two groups and grouping was done according to their collection regions. The study demonstrates higher genetic variability among studied genotypes with the suitability of ISSR markers for phylogenetic analysis of S. asoca genotypes. The findings of present investigation shall be helpful in the preparation of strategies for the management of Ashoka. The grouping of genotypes on the basis of their collection areas opens the window to perform future research for variations in the levels of bioactive compounds present in geographically distinct S. asoca genotypes of India.
Genetic Diversity, Genotypes, Dendrogram, ISSR, Conservation, Markers
Yadav S. S, Waoo A. A, Tiwari S. K. Identification of Genetic Relationship Among Saraca asoca Genotypes Using Inter Simple Sequence Repeat Markers. Biosc.Biotech.Res.Comm. 2020;13(4).
Yadav S. S, Waoo A. A, Tiwari S. K. Identification of Genetic Relationship Among Saraca asoca Genotypes Using Inter Simple Sequence Repeat Markers. Biosc.Biotech.Res.Comm. 2020;13(4). Available from: https://bit.ly/3nObpT1
Copyright © Yadav et al., This is an Open Access Article distributed under the Terms of the Creative Commons Attribution License (CC-BY) https://creativecommons.org/licenses/by/4.0/, which permits unrestricted use distribution and reproduction in any medium, provide the original author and source are credited.
INTRODUCTION
Saraca asoca (Roxb.) De Wilde, (Family: Caesalpiniaceae), a slow-growing climax forest tree species, is immensely valued for its medicinal properties. S. asoca is facing a problem of depletion from its innate habitation in India. It is now categorized as ‘vulnerable’ and considered ‘red listed’ by the International Union for Conservation of Nature (IUCN) (Senapati et al. 2012; Mohan et al. 2017). Due to historical evidences S. asoca, is generally acknowledged as the ‘Ashoka’. This species is found in evergreen forests as well as some other parts of India according to the suitability of climatic conditions. In terms of therapeutic importance of S. asoca, different plant parts such as bark, leaves, flowers and seeds have been found better and these plant parts are currently in use for the production of various medicines (Hegde et al. 2017a). The bark of S. asoca has proven its values better among all parts of the plant. It has been used in the preparation of such formulations which are generally used to treat gynecological disorders like menorrhagia, bleeding haemorrhoids and disorders associated with the menstrual cycle (Singh et al. 2015). Due to higher therapeutic values the plant is extensively exploited in the legal as well as illegal herbal drug trades (Hegde et al. 2017b; Hegde et al. 2018b; Yadav et al 2019).
Due to the increasing demand of this plant species, the enormous harvesting has pushed the species under ‘red listed’ category in a short period (Hegde et al. 2018b; Yadav et al 2019). Accurate identification of inherent variability in a rare and endangered plant species is necessary (Nongrum et al. 2012) to save them in their natural environment (Iranjo et al. 2016). Molecular characterization of genotypes of any endangered species collected from wild sources makes a platform to build conservation policies for future deployment of targeted species. Assessment of genetic diversity needs authentic tools which may not be influenced by environmental conditions. Molecular markers are DNA based tools for the analysis of genetic variations within and among the species (Tripathi et al. 2012; Tripathi and Khare, 2016; Hegde et al. 2018a). Hence, the application of molecular markers is important to generate diversity-based data for accurate execution of conservation strategies (Thakur et al 2019).
Molecular markers have also been applied in the field of crop genetics as well as hybridization programmes to transfer targeted gene/s (Kachare et al. 2019; Tiwari et al. 2019). Among all molecular markers inter simple sequence repeats (ISSRs) have proved their ability to characterize various medicinal plant species (Hadipour et al. 2020). These markers are able to generate large amount of precious information on the characterization of plant populations despite having few drawbacks such as reproducibility of bands. However, ISSR markers have been enormously applied to characterize various medicinal plant species due to the easy application and non requirement of prior acquaintance of DNA sequences (Tripathi et al. 2013; Sharma et al. 2016; Hadipour et al. 2020). The genetic diversity analysis among S. asoca genotypes collected from different locations including Central India has not been carried out earlier (Hadipour et al. 2020).
Among dominant molecular markers, the superiority of ISSRs has been proved previously in different medicinally important plant species and there are very few reports on the application of ISSR markers in S. asoca (Hadipour et al. 2020). So, the objective of the present study was to assess the genetic variability present among collected Indian S. asoca genotypes on the basis of ISSR fingerprinting technique.
MATERIAL AND METHODS
Initially an extensive survey was carried out to identify the location of India where Saraca asoca found. These locations were considered as potential pockets and geographical parameters (latitude and longitude) of such locations were noted down in the research notebook. For plant material, a total 15 plants of Saraca asoca (Table 1) were collected from potential pockets of India to assess genetic variability among them for the future conservation purposes.
Table 1. Details of collection site of Saraca asoca genotypes
| S. no. | Place of collection | Sample code | State | Latitude | Longitude |
| 1 | Kodaikanal-1 | SA1 | Tamilnadu | 10° 15′ N | 77 ° 33′ E |
| 2 | Kodaikanal-2 | SA2 | Tamilnadu | 10° 15′ N | 77 ° 33′ E |
| 3 | Melpallum | SA3 | Tamilnadu | 10° 20′ N | 77 ° 33′ E |
| 4 | Palani | SA4 | Tamilnadu | 10° 26′ N | 77 ° 29′ E |
| 5 | Satyamagalum-1 | SA5 | Tamilnadu | 11° 29′ N | 77 ° 13′ E |
| 6 | Satyamagalum-2 | SA6 | Tamilnadu | 11° 29′ N | 77 ° 13′ E |
| 7 | Satyamagalum-3 | SA7 | Tamilnadu | 11° 29′ N | 77 ° 13′ E |
| 8 | Satyamagalum-4 | SA8 | Tamilnadu | 11° 29′ N | 77 ° 13′ E |
| 9 | Munnar road forest area-1 | SA9 | Kerala | 10° 11′ N | 77 ° 15′ E |
| 10 | Munnar road forest area-2 | SA10 | Kerala | 10° 11′ 11N | 77 ° 15′ E |
| 11 | Gopal swami hill | SA11 | Karnataka | 11.12° 42′ N | 76 ° 36′ E |
| 12 | Borivali forest area | SA12 | Maharashtra | 19 ° 14′ N | 72° 50′ E |
| 13 | Vasco | SA13 | Goa | 15 ° 24′ N | 73 ° 50′ E |
| 14 | Veterinary College, Jabalpur | SA14 | Madhya Pradesh | 23 ° 06′ N | 79° 55′ E |
| 15 | Gwarighat, Jabalpur | SA15 | Madhya Pradesh | 23 ° 06′ N | 79° 55′ E |
For DNA extraction, genomic DNA of each collected plant of S. asoca was isolated using protocol standardized by Doyle and Doyle (1990). Extracted DNA was purified to remove the impurities like RNA, proteins and polysaccharides. Purity of DNA was checked by taking the ratio of Optical Density (O.D.) using UV- Spectrophotometer at 260 nm to that of 280 nm. The quality of DNA was also checked by horizontal submarine gel electrophoresis on 0.8% agarose gel. The quantity, quality and integrity of isolated DNA were also checked by gel electrophoresis.
PCR amplification and electrophoresis: For molecular diversity analysis initially a total of 40 ISSR primers were screened to check amplification efficiency with randomly selected four DNA samples as template. Amplification of ISSR fragments was performed in 25 μL volumes containing 30 ng genomic DNA, 10 pmol primer (IDT, India), 200 μM of each dNTP and 1 unit of Taq DNA polymerase (Promega) in PCR buffer supplied (TrisHCl, pH 9.0; 15 mM MgCl2). The amplification reaction consisted of an initial denaturation at 94 °C for 5 min followed by 40 cycles of denaturation at 94 °C for 60 s, annealing at 50 °C for 60 s, and extension at 72 °C for 1 min with final extension at 72 °C for 7 min.
ISSR amplifications were performed in thermal cycler (eppendrof realplex) thermal cycler. The amplified PCR products were visualized through gel electrophoresis (GENETIX GX606C) on 1-5% w/v agarose gels in 1X TAE buffer (40 mmol/L Tris, 20 mmol/L acetic acid, 1 mmol/L EDTA) using ethidium bromide as the staining dye, for 2.0 h at a constant voltage of 70 V. The agarose gels were visualized and documented under UV light using a gel documentation system with (geneview W645SC). Reproducibility of bands produced by each primer was confirmed with twice amplification of same bands with each primer and consequently, the steady and reproducible bands were scored for further data analysis.
For data analysis, band scoring was performed on the basis of their presence and absence. The presence of band was denoted by ‘1’ while the absence of band was denoted by ‘0’. After scoring the binary data matrix was prepared using excel sheet. A dendrogdocumentation system withram was constructed by using Unweighted Pair Group Method with Arithmetic average (UPGMA) cluster analysis based on the matrix of Jaccard’s similarity coefficient with NTSYS-pc (Rohlf, 2000) ver.2.1 to show a representation of genetic relationships among the studied S. asoca genotypes.
RESULTS AND DISCUSSION
Inter simple sequence repeat markers technology is one of the easiest and cheaper than other techniques available for DNA fingerprinting. Because of mentioned properties, this technique is also considered to be better than other random fingerprinting methods (Tripathi et al. 2012). Genetic diversity assessment is very imperative for the maintenance of plant genetic resources in their innate habitat. Initially, the amplification of isolated template DNA samples of S. asoca plants was performed with 40 ISSR primers (Tripathi et al. 2018; Liu et al. 2020).
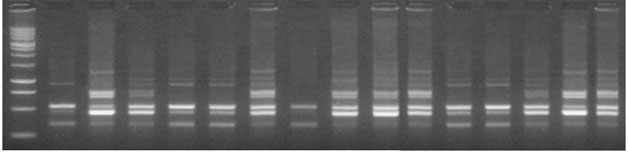
Among them only twenty eight primers (Table 2) have been found to be consistent with clear bands and were further selected for the study. Electrophoretic banding pattern of UBC 880 primer is illustrated in fig. 1. During the present study, total 149 bands were amplified and out of these 133 bands were found to be polymorphic and 16 bands were monomorphic. Monomorphic bands are those which are present in all individuals, polymorphic are present in one or more but not all individuals (Mehetre et al. 2004).The average numbers of total band was 5.321 while average numbers of polymorphic bands was 4.75. The number of bands produced per primer ranged from 2 (IUF022) to 8 (UBC 880).
Figure 1: Electrophoretic banding pattern of ISSR primer UBC 880 with 15 S. asoca genotypes
Table 2. Details of ISSR primers with banding profile used in the present study
| S. no. | Primer | Sequence 5’-3’ | TNB | PB | MB | PP |
| 1 | UBC 807 | AGAGAGAGAGAGAGAGT | 7 | 6 | 1 | 85.7 |
| 2 | UBC 808 | AGA GAG AGA GAG AGA GC | 6 | 5 | 1 | 83.3 |
| 3 | UBC 809 | AGA GAG AGA GAG AGA GG | 7 | 6 | 1 | 85.7 |
| 4 | UBC 810 | GAG AGA GAG AGA GAG AT | 7 | 6 | 1 | 85.7 |
| 5 | UBC 811 | GAG AGA GAG AGA GAG AC | 5 | 5 | 0 | 100 |
| 6 | UBC 812 | GAG AGA GAG AGA GAG AA | 4 | 4 | 0 | 100 |
| 7 | UBC 814 | CTC TCT CTC TCT CTC TA | 6 | 6 | 0 | 100 |
| 8 | UBC 816 | CAC ACA CAC ACA CAC AT | 6 | 6 | 0 | 100 |
| 9 | UBC 817 | CAC ACA CAC ACA CAC AA | 4 | 4 | 0 | 100 |
| 10 | UBC 818 | CAC ACA CAC ACA CAC AG | 6 | 6 | 0 | 100 |
| 11 | UBC 820 | GTGTGTGTGTGTGTGTC | 5 | 4 | 1 | 80.0 |
| 12 | UBC 830 | TGTGTGTGTGTGTGTGG | 4 | 4 | 0 | 100 |
| 13 | UBC 834 | AGAGAGAGAGAGAGAGYT | 6 | 5 | 1 | 83.3 |
| 14 | UBC 840 | GAGAGAGAGAGAGAGAYT | 5 | 5 | 0 | 100 |
| 15 | UBC 843 | CTCTCTCTCTCTCTCTRA | 6 | 6 | 0 | 100 |
| 16 | UBC 844 | CTCTCTCTCTCTCTCTRC | 5 | 5 | 0 | 100 |
| 17 | UBC 851 | GTGTGTGTGTGTGTGTYG | 6 | 5 | 1 | 83.3 |
| 18 | UBC 857 | ACACACACACACACACYG | 6 | 4 | 2 | 66.6 |
| 19 | UBC 860 | TGTGTGTGTGTGTGTGRA | 5 | 5 | 0 | 100 |
| 20 | UBC 862 | AGCAGCAGCAGCAGCAGC | 4 | 3 | 1 | 75.0 |
| 21 | UBC 868 | GAAGAAGAAGAAGAAGAA | 4 | 3 | 1 | 75.0 |
| 22 | UBC 880 | GGAGAGGAGAGGAGA | 8 | 8 | 0 | 100 |
| 23 | UBC 881 | GGGTGGGGTGGGGTG | 4 | 4 | 0 | 100 |
| 24 | IUF016 | ACTGACTGACTGACTG | 4 | 3 | 1 | 75.0 |
| 25 | IUF017 | GACACGACACGACACGACAC | 6 | 6 | 0 | 100 |
| 26 | IUF019 | AGAGAGAGAGAGAGAGAGAGG | 5 | 3 | 2 | 60.0 |
| 27 | IUF021 | AGAGAGAGAGAGAGAGAGAGC | 6 | 4 | 2 | 66.6 |
| 28 | IUF022 | AGAGAGAGAGAGAGAGAGAGT | 2 | 2 | 0 | 100 |
| Total | 149 | 133 | 16 | — | ||
| Average | 5.321 | 4.75 | 0.57 | 89.47 | ||
TNB-Total numbers of bands, PB-Polymorphic band, MB-Monomorphic bands, PP-Percentage polymorphism, Y= (C or T), R= (A or G)
Among all studied markers the highest percentage (100%) of polymorphism was demonstrated by fifteen markers however thirteen markers had less polymorphism comparatively. The lowest polymorphism (60%) was demonstrated by marker IUF019. The average percentage of polymorphism was 89.47%. However, only 43% variations have been reported previously by Hegde et al. (2018a) among S. asoca genotypes while studying genetic diversity analysis among different populations with the use of ISSR markers (Hegde et al. 2018a).
Cluster analysis was performed on the basis of similarity co-efficient generated from ISSR profiles. The cluster analysis grouped all the S. asoca genotypes under study in two groups i.e. group A and group B (Fig. 2). Group A is a major cluster consisting 13 S. asoca genotypes. Group A was further divided into two sub groups C and D. Sub group C consisted only two genotypes coded as SA-9 and SA-10. Both of these genotypes were collected from Kerala. Sub group D contained 11 genotypes of S. asoca. Among the genotypes clustered together in sub group D a total of 8 genotypes of S. asoca collected from different locations of Tamilnadu. Remaining 3 three genotypes in sub group D were from Karnataka, Maharashtra and Goa (Hegde et al. 2018a).
Figure 2: Inter Simple Sequence Repeat markers based dendrogram representing genetic relationship among 15 Saraca asoca genotypes
The clustering of genotypes in sub group D demonstrated higher similarity among the genotypes collected from Tamilnadu. However, the genotypes collected from Maharashtra, Goa and Karnataka showed genetic distance from Tamilnadu genotypes and were clustered distantly. Group B contained only two genotypes collected from Jabalpur, Madhya Pradesh. Both of these genotypes had higher resemblance with each other and clustered together. However, these both of the genotypes had high level of genetic dissimilarity with other studied genotypes so, these both were clustered distantly.
The grouping of the genotypes demonstrated genetic similarity between the genotypes collected from same geographical locations. Only few previous reports are available on clustering information of S. asoca genotypes collected from different locations with the use of ISSR markers. Hegde et al. (2018a) found similar clustering pattern during their study on S. asoca. In their report genotypes under cultivation were grouped separately from the individuals of wild genotypes (Hegde et al. 2018a).
The findings of the current investigation revealed the successful utilization of ISSR markers for the assessment of available variability among 15 S. asoca genotypes at molecular level. Idrees and Irshad (2014) stated the polymorphism showed by molecular markers may be due to change or mutation in targeted loci or variation of nucleotide. These alterations are responsible for the presence of genetic variability between and among individuals. In recent studies, ISSR have been applied for the assessment of inherent multiplicity in diverse endangered species medicinal plants (Hamouda 2019; Hadipour et al. 2020).
CONCLUSION
In conclusion, the present study demonstrated the applicability of ISSR markers in molecular variability assessment of 15 Indian S. asoca genotypes collected from various locations of the country. ISSR markers were found to be suitable to group the studied genotypes according to their collection sites. S. asoca genotypes collected from Kerala as well as Madhya Pradesh showed higher genetic distance from other genotypes. However, genotypes of Tamilnadu state had close similarity and they grouped together. Data generated in the present study will provide a base to develop conservation strategies for medicinally significant S. asoca species which has been already pushed into ‘red listed’ category.
ACKNOWLEDGEMENTS
Author are thankful to The Director, State Forest Research Institute Jabalpur for providing laboratory facilities to conduct the experiment and author also thankful to Department of Biotechnology, AKS University, Satna for providing research platform.
Conflict of interest: Authors declare no conflict of interest.
REFERENCES
Doyle JJ and Doyle JL (1990) Isolation of plant DNA from fresh tissue. Focus, 12, 3–15.
Hadipour M, Kazemitabar SK, Yaghini H and Dayani S (2020) Genetic diversity and species differentiation of medicinal plant Persian Poppy (Papaver bracteatum L.) using AFLP and ISSR markers. https://doi.org/10.1016/j.egg.2020.100058.
Hamouda M (2019) Molecular analysis of genetic diversity in population of Silybum marianum (L.) Gaertn in Egypt. Journal, Genetic Engineering & Biotechnology, 17(1),12
DOI: 10.1186/s43141-019-0011-6.
Hegde S, Hegde HV, Jalalpure SS, Peram MR, Pai SR and Roy S (2017a) Resolving identification issues of Saraca asoca from its adulterant and commercial samples using phytochemical markers. Pharmacognosy Magazine, 13(Suppl 2), S266.
Hegde S, Pai SR, and Roy S (2017b) Combination of DNA isolation and RP-HPLCanalysis method for bark samples of Saraca asoca and its adulterant.3Biotech, 7(3), 208.
Hegde S, Pai SR, Bhagwat RM, Saini A, Rathore PK, Jalalpure SS, Hegde HV, Sugunan AP, Gupta VS, Kholkute SD and Roy S (2018a) Genetic and phytochemical investigations for understanding population variability of the medicinally important tree Saraca asoca to help develop conservation strategies. Phytochemistry, 156, 43–54.
Hegde S, Saini A, Hegde HV, Kholkute SD and Roy S (2018b) Molecular identification of Saraca asoca from its substituents and adulterants. 3Biotech, 8(3), 161.
Idrees M and Irshad M (2014) Molecular markers in plants for analysis of genetic diversity: A review. Eurasian Academic Research 2, 1513–1540.
Iranjo P, NabatiAhmadi D, Sorkheh K, Memeari HR and Ercisli S (2016). Genetic diversity and phylogenetic relationships between and within wild Pistacia species populations and implications for its conservation. Journal of Forestry Research, 27(3), 685–697.
Kachare S, Tiwari S, Tripathi N and Thakur VV (2019) Assessment of genetic diversity of soybean (Glycine max (L.) Merr.) genotypes using qualitative traits and microsatellite makers. Agricultural Research, DOI: 10.1007/s40003-019-00412-y.
Liu X, Du J, Khan M, Cheng J, Wei C, Mei Z, Chen H, He T and Fu J (2020) Analysis of genetic diversity and similarities between different Lycium varieties based on ISSR analysis and RAMP‑PCR markers. World Academic Science Journal 2, 83-90.
Mehetre SS, Gomes M and Eapen S (2004) RAPD analysis of hybrid nature of the offspring of Gossypium hirsutum x G. raimondii. Current Science 84, 24–28.
Mohan C, Reddy MS, Naresh B, Kumar SM, Fatima S, Manzelat B and Cherku PD (2017) RAPD studies of Saraca asoca by fluorescent-labeled primers and development of micropropagation protocol for its conservation. International Journal of Applied Agricultural Research, 12(2), 137–151.
Nongrum I, Kumar S, Kumaria S and Tandon P (2012). Genetic variation and gene flow estimation of Nepenthes khasiana Hook. F – a threatened insectivorous plant of India as revealed by RAPD markers. Journal of Crop Science and Biotechnology, 15(2), 101–105.
Rohlf FJ (2002) NTSYS-pc: numerical taxonomy system ver.2.1.Exeter Publishing Ltd, Setauket.
Senapati SK, Das GK, Aparajita S and Rout, GR (2012). Assessment of genetic variability in the Asoka Tree of India. Biodiversity, 13(1), 16–23.
Sharma P, Tiwari S, Tripathi N and Mehta AK (2016) Polymorphism analysis in advanced mutant population of oat (Avena sativa L.) using ISSR markers. Physiology and Molecular Biology of Plants 22,115-120.
Singh S, Krishna AT, Kamalraj S, Kuriakose GC, Valayil JM and Jayabaskaran C (2015) Phytomedicinal importance of Saraca asoca (Ashoka): an exciting past, an emerging present and a promising future. Current Science, 109(10), 1790–1801.
Thakur VV, Tiwari S, Tripathi N and Tiwari G (2019) Molecular identification of medicinal plants with amplicon length polymorphism using universal DNA barcodes of the atpF-atpH, trnL and trnH-psbA region. 3Biotech 9, 188. https:// doi.org/10.1007/s13205-019-1724-6.
Tiwari S, Tripathi N, Tsuji K and Tantwai K (2019) Genetic diversity and population structure of Indian soybean (Glycine max (L.) Merr.) as revealed by microsatellite markers. Physiology and Molecular Biology of Plants 25, 953-964.
Tripathi N and Khare D (2016) Molecular approaches for genetic improvement of seed quality and characterization of genetic diversity in soybean: a critical review. Biotechnology Letters 38, 1645-1654.
Tripathi N, Saini N and Tiwari S (2013) Morphological and molecular characterization of endangered medicinal plant species Coleus forskohlii collected from central India. Journal of Crop Science and Biotechnology 16(4), 253- 261.
Tripathi N, Saini N, Kumar S and Tiwari S (2012) Assessment of Genetic Diversity among Withaniasomnifera Collected from Central India using RAPD and ISSR Analysis. Journal of Medicinal and Aromatic Plant Science and Biotechnology 6 (1): 133-139
Tripathi N, Shrivastava D, Mir BA, Kumar S, Govil S, Vahedi M and Bisen PS (2018) Metabolomic and biotechnological approaches to determine therapeutic potential of Withania somnifera (L.) Dunal: A review. Phytomedicine 50,127-136.
Yadav SS, Woo AA and Choure K (2019) Review of medicinally important plant species Saraca asoca (ROXB.). International Journal of Advanced Research. 7, 154-166.