1Department of Biology, College of Science, University of Jeddah, Jeddah, Saudi Arabia.
2Department of Agricultural Botany, Faculty of Agriculture, Suez Canal University, Ismalia, Egypt.
3Botany and Microbiology Department, Faculty of Science, Tanta University, Tanta, Egypt.
Corresponding author email: waalshehri@uj.edu.sa
Article Publishing History
Received: 28/06/2021
Accepted After Revision: 09/09/2021
Selection of appropriate strains of microalgae that work well in local conditions is important for a establish algae-based production system. The general aim of this study was to isolate and identify native microalgae species for exploring its further potential applications. To achieve this aim, 25 samples were collected from different locations of western region of Saudi Arabia. Standard isolation and purification techniques were applied In vitro to obtain axenic cultures. Among the best performing isolates, seven predominant strains were chosen for characterization based on morphological and molecular features. Morphological observations and molecular markers analysis using internal transcribed spacer sequence (ITS) were performed.
Moreover, phylogenetic relationship of these strains was constructed. According to the DNA sequence analysis of the seven isolates, they were belonged to six genera of Chlamydomonas, Dunaliella, Chlorococcum, Graesiella, Coelastrella and Chlorella. Screening the growth rates of all strains showed that Chlorella sorokiniana had the highest growth rate (0.180 day -1) and biomass productivity (150 mg. L-1.day -1). Whereas other strains showed comparable growth rates under same growth conditions. This study found that Chlorella sorokiniana UJ as robust species which holds a great potential to be used in different commercial and environmental applications. In conclusion the identification of microalgae is considered key step in microalgae-based industry. This work screened microalgae strains isolated from the local environment of Saudi Arabia. The best performing algal strains were selected to be identified and characterized to discover strains that can be utilized for mass cultivation. This study successfully isolated and identified seven local strains, most of them are already known with high biomass productivity (fast growers) and are considered as good candidate to serve as platform for many further applications.
Characterization, Growth Rate, Identification, Microalgae Saudi Arabia.
Alshareef M, Elbeshehy E. K. F, Alshehri W. A, Omar H. H. Identification and Growth Characterization of Native Microalgae Isolated from Different Environments of Saudi Arabiaa. Biosc.Biotech.Res.Comm. 2021;14(3).
Alshareef M, Elbeshehy E. K. F, Alshehri W. A, Omar H. H. Identification and Growth Characterization of Native Microalgae Isolated from Different Environments of Saudi Arabiaa. Biosc.Biotech.Res.Comm. 2021;14(3). Available from: < href=”https://bit.ly/3iw3PfU“>https://bit.ly/3iw3PfU</a>
Copyright ©Alshareef et al., This is an open access article distributed under the terms of the Creative Commons Attribution License (CC-BY) https://creativecommns.org/licenses/by/4.0/, which permits unrestricted use distribution and reproduction in any medium, provide the original author and source are credited.
INTRODUCTION
Algae constitute a diverse group of photosynthetic organisms, which ranging from single cellular bodies to multicellular seaweeds, with a broad diversity in morphological, physiological and biochemical characteristics, distributed in almost all environments (Buijks, 2012). Using microalgae have attracted much attention in various industrial and environmental sectors such as human food, animal and aquaculture feed, pharmaceuticals, cosmetics, wastewater treatment, and bio-fertilizers (Olaizola, 2003; Draaisma et al., 2013).
Moreover, microalgae are considered as promising alternative biofuel feedstocks due to their rapid growth, high biomass productivity and their capability to grow under divers conditions (Wijffels and Barbosa, 2010; Elliott et al., 2012; Ratha and Prasanna, 2012; Ratha et al., 2012; Markou and Nerantzis, 2013).
Over the last decade, a number of microalgae have been cultivated on large scale to be used in industry because of their ability to produce valuable products (Olaizola, 2003; Markou and Nerantzis, 2013; Wijffels et al., 2013; Kaspar et al., 2014; Mulders et al., 2014; Ciurli et al., 2021). The common genera used are Spirulina and Chlorella as nutritional supplement, Haematococcus, to produce the antioxidant astaxanthin (Guerin et al., 2003; Yuan et al., 2011), and Dunaliella salina to produce carotenoids (Borowitzka, 1999; Hosseini Tafreshi and Shariati, 2009; Knothe, 2010; Camacho et al., 2019; Ciurli et al., 2021).
Due to these successful achievements of the using algae, the unexploited and undiscovered microalgae offer great possibilities for future. Until now, the actual number of microalgae species are still unknown, it is estimated that there are between 200,000 and 800,000 species (De Clerck et al., 2013; Lizzul et al., 2018; Camacho et al., 2019; Yin et al., 2020; Ciurli et al., 2021). Only 50,000 species have been defined and characterized (Yin et al., 2020). New genera and species are being discovered very consistently indicating the presences of large portion of undescribed species that exist (De Clerck et al., 2013).
Saudi Arabia could be a good source of algal biodiversity due to variation of geographical and environmental nature. The capability of microalgae to grow in the local environmental conditions is an important prerequisite toward successful cultivation of microalgae-based for industrial production. Therefore, there is a need for a proper identification and characterization of local species. Microalgae are usually identified based on their morphological features.
However, environmental factors may cause some of the phenotypic plasticity, therefore morphological identification could be insufficient and misleading (Hoshina et al., 2010; Gour et al., 2016). A new classification based on the ultra-structure of the basal body in the flagellation cells and cytokinesis during mitosis has been suggested. Nevertheless, these principles are difficult to practice especially by non-taxonomists (Gour et al., 2016). The presence of molecular-based techniques such as polymerase chain reaction (PCR) and sequencing have improved such studies and helped in demonstrating evolutional relationships between different organisms and species.
Molecular based techniques are usually recommended to confirm morphological classifications (Chung et al., 2018). Nowadays, internal transcribed spacer sequence (ITS) is considered a very powerful and helpful tool to discriminate between microalgae at the genus and species level (An et al., 1999; Van Hannen et al., 2002; Coleman, 2003, 2009; Hegewald et al., 2005, 2010; Jeon and Hegewald, 2006; Schultz et al., 2006; Schultz and Wolf, 2009; Hegewald et al., 2013; Lizzul et al., 2018; Wang et al., 2019; Goecke et al., 2020; Karm and Dwaish, 2021).
Thus, in current study, amplification and sequencing of internal transcribed spacer sequence (ITS 1 and ITS 4) was chosen as molecular markers to confirm the primary morphological identification of isolated strains, compared with other known sequences of species from public databases. The results of these comparisons were represented in a phylogenetic tree. This sort of studies is limited in Saudi Arabia and therefore this work will contribute in the field of discovering and exploiting microalgae from the local environment.
MATERIAL AND METHODS
Sampling, isolation and purification of microalgae: Twenty-five samples were collected through duration (Jan-April, 2018) from different environments of western region of Saudi Arabia (Table 1). Once the samples were collected and transferred to the laboratory, they were exposed to light, enriched with BG-11 medium and incubated for few days (Rippka et al., 1979). In order to obtain an axenic culture, the basic microbiological techniques for isolation and purification were used (serial dilution in liquid media and streak plate method). In the serial dilution method, a series of test tubes with 9 ml of sterilized distilled water were prepared. One ml of the mixed enriched sample was taken, diluted in the first test tube (10−1) and mixed. Next, 1 ml was taken from the first dilution and transferred to the second test tube (10−2), this process was repeated until reaching to dilution of (10-6).
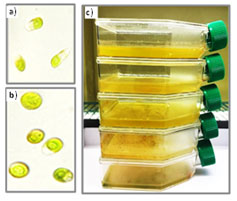
Serial dilution increases the chance of getting individual colonies. Then, each diluted tube was cultured in both liquid and semi-solid agar plates media, incubated in controlled conditions at 22 ± 1°C, exposed to continues light using LED fluorescent tube of intensity 2000 LUX 28 μmol.m-2. s-1 (Figure 1a). Further purification was achieved by consecutive streaking on semi-solid BG-11 agar (Figure 1b). This process was repeated several times until axenic culture was obtained. To ensure purity, the cultures were regularly monitored using light microscope. Subculturing was performed every 2 weeks. Axenic culture was preserved in cell culture flasks (Figure 1c).
Figure 1: Isolation and purification of algae. a) Algae stock cultures, b) Streaking purification of the microalgae on semi-solid BG-11 agar plates, c) Axenic culture preserved in cell culture flasks.

Growth of microalgae isolates in different media: Algae isolates were cultivated on different growth media, Kuhl (SAG Göttingen, 2013), BG-11 (Rippka et al., 1979), F2 (Guillard and Ryther, 1962) and Johnson’s (Johnson et al., 1968). The cultures were aerated and incubated under controlled condition mentioned previously. The growth of algae was estimated and determined as shown in (Table1).
Morphological identification: To identify morphological characterization of isolated strain, cell shape and arrangement were documented using light microscope (BX51; Olympus, Tokyo, Japan) equipped with a built-in digital camera, and microphotographs were processed with cell Sens Standard program. Morphological identification was determined according to Sime (2004) and Serediak and Huynh (2011).
Genomic DNA Isolation and PCR amplification of ITS Regions: For DNA isolation, 50 ml of each culture were harvested using centrifuge at 5000x g for 15 minutes, the genomic DNA (gDNA) then extracted using Qiagen kit following the manufacturer’s instructions. The extracted gDNA was confirmed by agarose gel electrophoresis (1%) stained with ethidium bromide and visualized under ultraviolet light (UV). The extracted DNA was kept at –20 °C till using as PCR template. To amplify the ITS gene, the universal oligonucleotide primer set described in Van Hannen et al., (2002) was used. The sequences of these primers are: FW primer ITS1: (5′- TCCGTAGGTGAACCTGCGG -3′) RE primer ITS4: (5′- TCCTCCGCTTATTGATATGC -3′)
The PCR reaction was performed in a total volume of 25 ml by mixing up the following reagents: 12.5 μL master mix (2x), 8.5 μL dH2O, 1 ml from each forward primer (FW) and reverse primer (RE) (10 pmoles) and 2 ml of genomic DNA. The PCR reaction was carried on at the thermocycler (Bibby Scientific, UK) using the following conditions: 5 minutes at 94 °C for the initial denaturation followed by 35 cycles of denaturation at 94 °C for 1 minute, annealing at 60 °C for 1 minute, extension at 72 °C for 2 minutes, followed by a final extension step at 72 °C of 10 minutes. Then the PCR products (860 bp) were resolved in 1.5 % agarose gel stained with ethidium bromide and visualized using UV light detector system. To perform DNA sequencing, the PCR products were shipped to (Macrogen, Seoul, Korea) to perform DNA sequencing reaction using Sanger sequencing methods.
Sequence analyses of ITS region: In order to identify the isolated strains, the obtained sequences were analyzed by searching for homology in the National Center for Biotechnology Information (NCBI) database using BLAST tool. All sequences were submitted to GenBank under the number SUB9546968. The phylogenetic tree of identified microalgae was constructed using the neighbor-joining (NJ) method (Ratha et al., 2012) of Mega 5 software (Draaisma et al., 2013). A bootstrap analysis of 1000 replication was used to test the degree of support of the branches produced by NJ analysis (Figure 2).
Growth characteristics: To study algal growth, four liters of BG-11 medium (except for S2 and S6 Jonson medium) was inoculated with active 7 days old inocula and incubated under controlled conditions as described above. Culture’s densities were adjusted to be (OD680 = 0.25) in zero day for all strains. Optical densities were measured on regular basis using spectrophotometer (T60 UV–visible spectrophotometer, PG instruments, UK). To determine the dry weight of the biomass, 60 ml of each culture were centrifuged at 4000×g for 10 minutes and the washed several times with distilled water to remove residual salts. Then, the pellets dried at 60 °C oven for 48 h, weighed. The specific growth rate (μ) of each strain was calculated using equation below (Gill et al., 2016) :
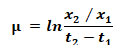
where x1 is weight of dry biomass at the beginning of the selected time interval, x2 is weight of dry biomass at the end of the selected time interval, (t2–t1) is the selected time for the determination of dry biomass. Biomass productivity (Pdwt) was expressed as dry biomass produced per litter per day (mg. L -1.day-1) during the exponential growth phase (Song et al., 2013) according to the following equation:
![]()
RESULTS AND DISCUSSION
The pre-enrichment step allowed strains with high growth performance to compete with other weak strains. Therefore, this step is important to screen the best candidate strain for further scale up cultivation. After two weeks of initial incubation, most isolates were showed growth in liquid media. The growth performance was noticed and reported as ++++, +++, ++, + or – (Table 1). It was clear that the maximum growth was mostly observed in BG-11 medium except for (S2 and S6) were they had the highest growth (++++) in Johnson’s medium.
Samples with good growth were examined under light microscope to determine the type of microalgae present in them. Even though sub-culturing was done with extreme care, in some samples, especially when they were grown in BG-11 medium, protozoa and rotifers prey on microalgae rapidly and could not be processed or rescued, because BG-11 medium was improved microscopic grazer growth along with microalgae. In case of marine-origin samples, the growth was very weak, thus, those cultures were discarded.
Microscopic investigation of several samples during the screening process revealed the presence of flagellated, coccoid and filamentous algae. In some cases of mixed culture with filamentous algae, the growth of filamentous algae were very dense and no single cell strains could be purified. In addition, filamentous algae have sticky nature and very hard to handled, hence, they were excluded from selected collection. Also, it was noticed that S11 (obtained from 70 °C hot spring) requires at least 40°C incubation temperature to grow, therefore it is excluded too.
Based on the growth performance and microscopic examination, seven isolates (S1, S2, S3, S4, S5, S6, S7) were chosen for morphological and molecular identification. For subsequent sub-culturing rounds, most microalgae were showed growth after seven days in the agar surface, probably due to acclimatization in the new conditions. Whereas in case of S2 and S6 (which later defined as Dunaliella salina), they did not show growth on agar plates, however, they grew well in liquid media.
Table 1. Growth of the isolated microalgae on different media after 14 days. Excellent growth (++++), moderate growth (+++), poor growth (++), very poor growth (+), no growth (-).
| Isolate ID | Source | Habitat | Isolation location | Medium | |||
| Kuhl | BG-11 | F2 | Jonson | ||||
| S1 | Fresh | Agriculture soil | Al-Madina | + | ++++ | – | – |
| S2 | Fresh | Anthropogenic silt soil | Al-Baqi cemetery | – | ++ | – | ++++ |
| S3 | Fresh | Agriculture soil | Al-Madina | ++ | ++++ | – | – |
| S4 | Fresh | Stagnant water (effluent from air conditioner). | Jeddah | + | ++++ | – | – |
| S5 | Fresh | Agriculture soil | Al-Madina | + | ++++ | – | ++++ |
| S6 | Marine | Al Khumra salt marshes | Jeddah | + | + | ++ | ++++ |
| S7 | Fresh | Agriculture soil | Jeddah | + | ++++ | – | – |
| S8 | Marine | Sharm beach | Yanbu | – | – | + | – |
| S9 | Fresh | Asphalt surface | Jeddah | ++ | ++++ | + | – |
| S10 | Fresh | Agriculture soil from public walkway | Jeddah | – | ++++ | – | – |
| S11 | Fresh | Gomygah hot spring | Al-Lith | + | ++ | – | – |
| S12 | Fresh | Damp walls | Jeddah | – | ++++ | – | – |
| S13 | Fresh | Garden irrigation dripper | Jeddah | + | ++++ | – | – |
| S14 | Fresh | Birdbath | Jeddah | + | ++++ | – | – |
| S15 | Fresh | Water barrel | Jeddah | + | ++++ | – | – |
| S16 | Fresh | Planter | Jeddah | + | ++++ | – | – |
| S17 | Fresh | Fountain | Jeddah | + | +++ | – | – |
| S18 | Fresh | Roadside mud | Al-Lith | + | ++++ | – | – |
| S19 | Marine | Waterfront Corniche | Jeddah | – | – | + | – |
| S20 | Fresh | Agriculture soil | Jeddah | + | + | – | – |
| S21 | Fresh | Agriculture soil | Yanbu | + | ++++ | – | – |
| S22 | Fresh | Agriculture soil | Al-Taif | + | +++ | – | – |
| S23 | Fresh | Roadside soil | Al-Madina | + | ++++ | – | – |
| S24 | Fresh | Greenhouse hydroponics | Thuwal | + | ++++ | – | – |
| S25 | Marine | Thuwal beach | Thuwal | + | ++++ | – | – |
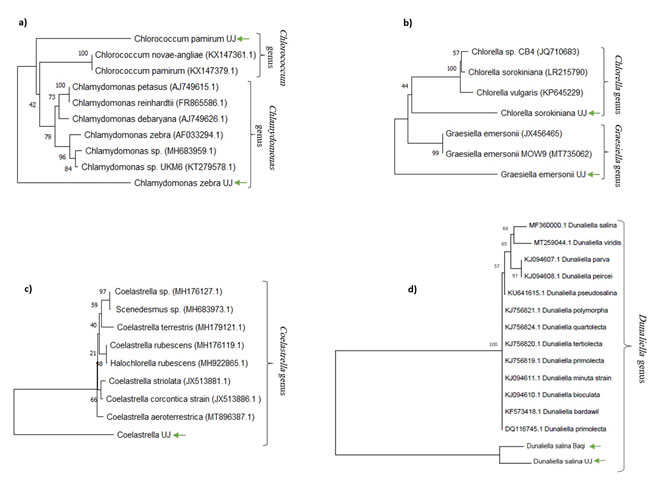
Morphological and molecular identification and characterization: In this study, seven strains were chosen to be identified and characterized. The selection criteria were focused on strains that could be easily cultivated. Based on morphological and reproductive features, all chosen strains were belonging to the phylum of Chlorophyta under the genera of Chlamydomonas, Dunaliella, Chlorococcum, Graesiella, Coelastrella, and Chlorella. To verify their taxonomical positions, the sequences of internal transcribed spacer (ITS) obtained from our samples were compared with the sequences available in the NCBI database, the results are summarized in (Table 2). Similar sequences were used to construct independent molecular phylogenetic trees. The reliability of the phylogenetic tree was evaluated using neighbor-joining analysis.
Table 2. Identification results of isolated stains according to the BLAST hits.
| E-value | Identity (%) | Nucleotide length | Coverage
(%) |
Name and accession of number of the most related strain in NCBI GenBank | Isolates | |
| 2E-114 | 90% | 333 | 98 % | AF033294.1 | Chlamydomonas zebra | S1 |
| 2E-99 | 98% | 221 | 99 % | MF360000.1 | Dunaliella salina | S2 |
| 7E-129 | 99% | 269 | 98 % | KX147379.1 | Chlorococcum pamirum | S3 |
| 3E-72 | 91% | 221 | 96 % | JX456465.1 | Graesiella emersonii | S4 |
| 0 | 100% | 493 | 100 % | MH176127.1 | Coelastrella sp. | S5 |
| 0 | 96% | 442 | 99 % | MF360000.1 | Dunaliella salina | S6 |
| 4E-75
|
95% | 221 | 85 % | LR215790.1 | Chlorella sorokiniana | S7 |
Strain (S1) Chlamydomonas zebra: Microscopic examination of S1 demonstrated green, motile, unicellular; oval shaped cells (4-5.5 μm long, 4-3 μm wide) showed morphology consistent with Chlamydomonas sp. features (Figure 3). Previous research described Chlamydomonas species as an ovoid-shaped unicellular, of (9-16 μm long, 5-12 μm wide) with two anterior isokont flagella and single cup-shaped chloroplast, single nucleus; two anterior contractile vacuoles. Reproduction by producing 2-8 zoospores; or by isogamous (Serediak and Huynh, 2011; Zhang et al., 2014).
The phylogenetic tree revealed that the sequence of S1 strain aligned with other strains of Chlamydomonas genus described in previous studies (Luo et al., 2010; Hoshina, 2014). According to the BLAST analysis results, the S1 is homologous and is closely related to the Chlamydomonas zebra. Therefore, the isolated S1 strain was given the name Chlamydomonas zebra UJ (Figure 2 a).
Figure 2: Microscopic photograph of Chlamydomonas zebra cells at a magnification of 400x.
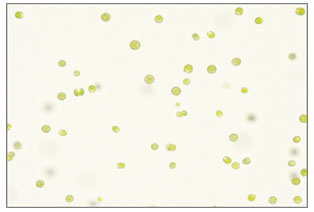
Figure : Neighbor-joining phylogenetic tree of ITS region sequences of isolated algae.
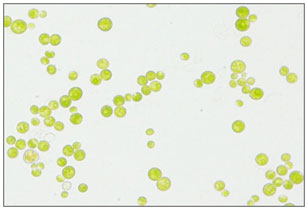
Strain (S3) Chlorococcum pamirum: Microscopic observation of S3 showed green spherical cells, vary in size (4.5-6 μm in diameter), solitary and form temporary groups (Figure 4). This morphology was similar to Chlorococcum sp. as mentioned in Feng et al., (2014), vegetative cells range in diameter from 5-16 μm. The cells have a distinct pyrenoid surrounded by a sheath of biplate starch and are uninucleate.
Reproduced asexually by zoospores and aplanospores or sexually by isogametes (Blackwell, Cox and Gilmour, 1991). Therefore, S3 was preliminarily hypothesized to belong to genus Chlorococcum sp. The BLAST analysis result supported our hypothesis, as shown in phylogenetic tree (Figure 2 a). The sequence of S3 was located in the same clade with other Chlorococcum species, and therefore, it was given the name of Chlorococcum pamirum UJ.
Figure 4: Microscopic photograph of Chlorococcum pamirum cells at a magnification of 400x.
Strain (S4) Graesiella emersonii: Under light microscope, the cells of S4 strain were giant (diameter ranging 9-22 μm), non- motile, unicellular, nearly globose to ellipsoidal shape with different sizes, enclosed by thick transparent sheath, and form auto-spores as a mean of asexual reproduction (Figure 5). These observations were similar to Graesiella emersonii features described by Nozaki et al., (1995) as a large globose to ellipsoidal cells (up to 5-17 µm in diameter), surrounded by double-layered cell wall. Graesiella emersonii also had a massive chloroplast containing a single pyrenoid and exhibiting several vacuoles and reproduce asexually by autospores. The molecular analysis of ITS regions of S4 showed a high similarity to Graesiella emersonii (Table 2) which supports the morphological characteristics; therefore, it was given the name of Graesiella emersonii UJ (Figure 2 b).
Figure 5: Microscopic photograph of Graesiella emersonii cells at a magnification of 400x.
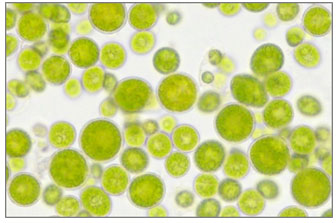
Strain (S5) Coelastrella sp.: Microscopic observation of isolated strain S5 shows there was similarity in morphological characteristic described in literature of Coelastrella sp. which is unicellular spherical cells, tend to aggerate in temporary groups (Figure 6 a,c). These characteristics were previously described in Coelastrella species (Wang et al., 2019; Goecke et al., 2020). Another important characteristic that has been reported in several Coelastrella species is the production of secondary pigments of carotenoids (Punčochářová and Kalina, 1981; Abe et al., 2007; Hu et al., 2013a; Kawasaki et al., 2020).
Changing of cell color in old and stressed cultures was noticed in S5 which indicating carotenoids production (Figure 6 b). Based on these finding, it is suggested that S5 to is a member of genus Coelastrella, and the molecular analysis supported this suggestion. Thus, the isolate S5 was given the name Coelastrella sp. UJ (Figure 2 c). Coelastrella species are widely distributed worldwide. It has been reported to be found in temporary waterbodies, birdbaths, and fountains (Neofotis et al., 2016). They found as single cell or in aggregations of few cells, these species are characterized by double layered cell wall, with distinct sculpture.
The inner layer composed of cellulose and an outer one composed from (sporopollenin) which is acetolysis-resistant material (Tschaikner and Kofler, 2008). Previous research noticed the presence of small thickenings at the poles of the cells a citriforme in addition of longitudinal ribs considered as an important character of Coelastrella species (Punčochářová and Kalina, 1981; Abe et al., 2007; Hu et al., 2013b; Kawasaki et al., 2020; Ciurli et al., 2021 ).
Coelastroideae subfamily members have been previously placed under family of Oocystaceae, Chlorellaceae, and Sctielloideae regarding to morphology and cellular structure (Kalina and Punčochářová, 1987). Later, the phylogenetic molecular studies suggested that Coelastroideae should be placed within the family Scenedesmaceae, order Sphaeropleales (Hanagata, 1998; Hegewald et al., 2010; Kaufnerová and Eliáš, 2013; Lee et al., 2016; Ancona-Canché et al., 2017). Nowadays, many species of this genus attract attention from researchers due to its ability of accumulation carotenoids and fatty acids, as well as for a potential use for bioremediation (Abe et al., 2007; Hu et al., 2013b; Kawasaki et al., 2013; Luo et al., 2016; Dimitrova et al., 2017; Thao et al., 2017; Goecke et al., 2020; Karm and Dwaish, 2021).
Figure 6: Microscopic photograph of Coelastrella sp. cells at a magnification of 400x. a) Cell’s aggregation, b) Orange cells indicating carotenoids production, c) Single spherical cell.
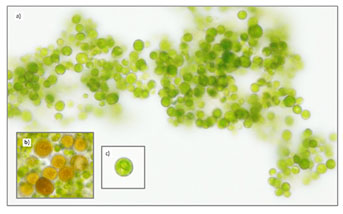
Strains (S2 and S6) Dunaliella salina: Although S2 and S6 obtained from different environments (anthropogenic silt soil and saltmarshes), respectively, both of them were shows similarity in morphological and physiological characteristic of Dunaliella species. Under microscope, ovoid motile cells with cup shaped chloroplast were observed (Figure 7 a,b ). It was also noticed that both strains change their cell color from green into orang under high light and salt stress (Figure 7 c).
Previous works described Dunaliella sp. characteristic as following: ovoid to spherical motile cell with two equal flagella and cup-shaped chloroplast. Depending on different environmental conditions, the size and shape of the cell can vary within a species (between 2 to 28 μm and in width between 1 to 15 μm) (Hosseini et al., 2009). Although, Dunaliella cells are naked, they are surrounded by mucilaginous substance (Ben Amtoz et al., 2009).
Furthermore, it is well known that many species of Dunaliella are halotolerant and capable to grow in environment of high salinities, they were previously isolated from Dead Sea, and the Great Salt Lake, USA (Oren, 2014). Dunaliella response to such environmental stress through over-accumulation of beta-carotene pigment which is responsible for turn cell color into orange (Polle et al., 2017).
Until now, 26 saltwater species and five freshwater species have been described for the genus Dunaliella. All freshwater species considered rare and its classification is still uncertain (Ben Amtoz et al., 2009; Gonzalez et al., 2001; Melkonian and Preisig, 1984). Molecular analysis of both isolated strains (S2 and S6) was confirmed the morphological and physiological findings. As our isolates were closely related to Dunaliella salina, they have been given the following names Dunaliella salina UJ and Dunaliella salina Baqi to distinguish its unique origin for further studies (Figure 2 d).
Figure 7: Light microscopic images (magnification of 400x) of Dunaliella salina. a) Strain isolated from saltmarshes, b) Strain isolated from anthropogenic silt soil, c) Orange color of Dunaliella salina stressed cultures.
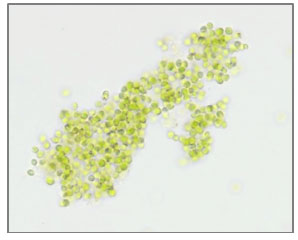
Strain (S7) Chlorella sorokiniana: Microscopic observation of S7 shows a small (diameter 2-2.5 μm) spherical unicellular, emerald-green color alga (Figure 8). These morphological characteristics are similar to the morphological characteristics of Chlorella genus (Krienitz and Bock, 2012; Lizzul et al., 2018). The molecular identification confirmed this finding, therefore, S7 was designated as Chlorella sorokiniana UJ (Figure 2 b). Chlorella genus members are widely distributed in different habitat due to their rapid growth (Lizzul et al., 2018), Chlorella species have been used as model organisms for photosynthesis studies and biotechnological applications for decades (Béchet et al., 2013; Lizzul et al., 2018).
To date there are more than 20 characterized Chlorella species (Furnas, 1990; Krienitz et al., 2015). In the past few years, the classification of the genus of Chlorella has received a lot of attention and many species within the genus were re-classified (Luo et al., 2010; Lemieux et al., 2014; Krienitz et al., 2015). Chlorella sorokiniana is a sub-species first isolated in 1953 by Sorokin, and believed to be a thermotolerant mutant of Chlorella pyrenoidosa (Sorokin and Myers, 1953; Kunz, 1972; Lizzul et al., 2018).
Later, this taxonomy was changed and re-classified C. sorokiniana as a separate species (Kessler, 1985; Dörr and Huss, 1990; Kessler and Huss, 1992). Furthermore, it is worth to mention that this sub-species is unique and robust alga due to its ability to thrive under harsh conditions such as high salinities and temperature up to 40 °C. Therefore, C. sorokiniana consider the subject of research in several major laboratories (de-Bashan et al., 2008; Lizzul et al., 2014, 2018; Krienitz et al., 2015; Neofotis et al., 2016; Jiang and Pei, 2021).
Figure 8: Microscopic photograph of Chlorella sorokiniana cells at a magnification of 400x.
Growth and biomass productivity: In general, algal growth can be monitored by determining the changes in biomass directly using cell count, or using other parameters such as chlorophyll a, optical density and dry weight (Richmond, 2003). The growth response of our isolates was measured using optical density at 680 nm on regular basis throughout the span of cultivation. The data obtained from OD showed similar trend for all species under same growth condition as presented in (Figure 9). The growth rate defined as the increasing of biomass over specific period of time (Richmond, 2003).
Figure 9: Growth curves of a) Chlamydomonas zebra UJ, b) Dunaliella salina UJ, c) Chlorococcum pamirum UJ, d) Graesiella emersonii UJ, e) Coelastrella sp. UJ, f) Dunaliella salina Baqi, g) Chlorella sorokiniana UJ, every point on the graph is showing the mean of three OD reading.
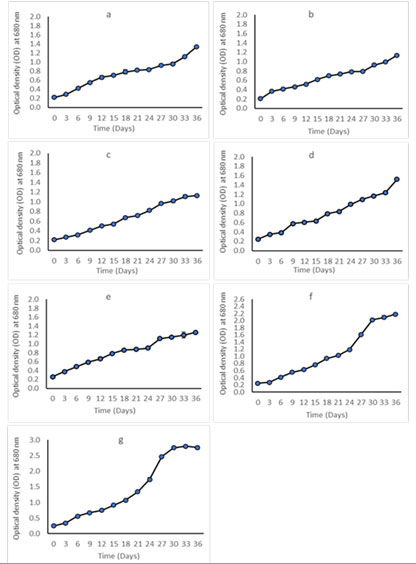
In this study, Chlorella sorokiniana had the highest growth rate followed by Dunaliella salina UJ and Ch–lamydomonas zebra. Whereas, Chlorococcum pamirums, Coelastrella sp. and Graesiella emersonii showed a comparable growth rate. The slowest growth rate was noted in Dunaliella salina Baqi. The growth parameters of all strains are shown in (Table 3).
Table 3. Algae species, culture medium, specific growth rate, and biomass productivity.
| Isolates | Algal species | Culture medium | Specific
growth rate (day-1) |
Biomass productivity
(mg. L-1.day-1) |
| S1 | Chlamydomonas zebra | BG-11 | 0.158 | 68.3 |
| S2 | Dunaliella salina | Johnson | 0.087 | 14.703 |
| S3 | Chlorococcum pamirum | BG-11 | 0.169 | 113.9 |
| S4 | Graesiella emersonii | BG-11 | 0.171 | 123.8 |
| S5 | Coelastrella sp. | BG-11 | 0.169 | 128.9 |
| S6 | Dunaliella salina | Johnson | 0.173 | 111 |
| S7 | Chlorella sorokiniana | BG-11 | 0.180 | 150.2 |
As growth rate measures the cellular response to nutrients and growth conditions, different algal strains grown under a variety of culture conditions gives a variable response depending on cultivation mode, types of media, temperature, light intensity, photoperiod, and supplying of CO2 (Enamala et al., 2018). Several studies performed previously to investigate the growth potential of different microalgae strains, however, these studies indicated that there is no standard species can be used to make precise comparisons, as well as the differences of cultivation conditions and methods (Richmond, 2003). Feng et al. (2014) found that growth rate of Chlorococcum pamirums was 1.88 day -1 which is slightly higher than our result. The growth rates of Dunaliella salina and Chlorella sorokiniana were found to be 0.16 -0.20 day -1 and 0.19 – 0.20 day -1 respectively (Pertumbuhan et al., 2017; Sajjadi et al., 2018; Khatoon et al., 2020; Karm and Dwaish, 2021).
Which are close to the growth rate obtained from our study (Table 3). Specific growth rate reflects the time required for cells to divide, however, some microalgae species grow by increasing their size rather than increasing their cell number (Zachleder et al., 2016) therefore, high growth rate does not necessary reflect high productivity. Thus, biomass productivity is usually used as a more reliable method to evaluate strain efficiency. Biomass productivity is generally calculated as the increase in biomass over a period of time. In this study, we compared the biomass productivity of all strains and found that Chlorella sorokiniana is the most productive strain, whereas Dunaliella salina Baqi was the least. In general, the productivity values obtained from this study (shown in Table 3) were approximately similar to the values mentioned in previous works (Khan et al., 2009; Rodolfi et al., 2009; Mata, Martins and Caetano, 2010; Park et al., 2012; Enamala et al., 2018; Sajjadi et al., 2018; Khatoon et al., 2020; Ciurli et al., 2021).
CONCLUSION
Identification of microalgae is considered a key step in microalgae-based industry. This work screened microalgae strains isolated from the local environment of Saudi Arabia. The best performing algal strains were selected to be identified and characterized to discover strains that can be utilized for mass cultivation. This study successfully isolated and identified seven local strains, most of them are already known with high biomass productivity (fast growers) and are considered as good candidate to serve as platform for many further applications.
Among these strains are Dunaliella and Coelastrella genera, these two genera were reported to be capable to accumulate carotenoids and thus they could be exploited for carotenoid production. We also identified strain of Chlorella sorokiniana which is currently one of the most promising algal species that can be used as biofuel feedstocks. The results of this study can greatly enrich our knowledge of microalgae biodiversity in Saudi Arabia. To the best of our knowledge, this study is one of few reports focus on exploring the local microalgae isolates. Integrating phyco-prospecting and characterizing native isolates could contribute in supporting algae-based industry for future.
ACKNOWLEDGEMENTS
We would like to thank King Fahd Medical Research Center (KFMRC), for providing facilities for conducting the research.
REFERENCES
Abe, K., Hattori, H. and Hirano, M. (2007). Accumulation and antioxidant activity of secondary carotenoids in the aerial microalga Coelastrella striolata var. multistriata. Food Chemistry, Vol 100(2), pp. 656–661. doi: 10.1016/j.foodchem.2005.10.026.
An, S. S., Friedl, T. and Hegewald, E. (1999). Phylogenetic relationships of Scenedesmus and Scenedesmus-like coccoid green algae as inferred from ITS-2 rDNA sequence comparisons. Plant Biology, Vol 1(4), pp. 418–428. doi: 10.1111/j.1438-8677.1999.tb00724.x.
Ancona-Canché, K. et al. (2017). Molecular phylogeny and morphologic data of strains of the genus Coelastrella (Chlorophyta, Scenedesmaceae) from a tropical region in North America (Yucatan Peninsula). Botanical Sciences, Vol 95(3), pp. 527–537. doi: 10.17129/botsci.1201.
Béchet, Q. et al. (2013). Outdoor cultivation of temperature-tolerant Chlorella sorokiniana in a column photobioreactor under low power-input. Biotechnology and Bioengineering. John Wiley and Sons Inc., Vol 110(1), pp. 118–126. doi: 10.1002/bit.24603.
Ben-Amotz, A., Polle, J. E.W. and Subba Rao, D.V. (2009). The Alga Dunaliella Biodiversity, Physiology, Genomics and Biotechnology, Science Publisher, New Hampshire.
Blackwell, J. R., Cox, E. J. and Gilmour, D. J. (1991). The morphology and taxonomy of Chlorococcum submarinum (Chlorococcales) isolated from a tidal rockpool. British Phycological Journal, Vol 26(2), pp. 133–139. doi: 10.1080/00071619100650101.
Borowitzka, M. A. (1999). Commercial production of microalgae: ponds, tanks, and fermenters. Progress in Industrial Microbiology, Vol 35(C), pp. 313–321. doi: 10.1016/S0079-6352(99)80123-4.
Buijks, J. (2012). Future of algae based biodiesel production in the Netherlands. pp. 48.
Camacho, F., Macedo, A. and Malcata, F. (2019). Potential industrial applications and commercialization of microalgae in the functional food and feed industries: A short review. Marine Drugs, Vol 17(6) pp .1-25. doi: 10.3390/md17060312.
Chung, T. Y. et al. (2018). Indole-3-acetic-acid-induced phenotypic plasticity in Desmodesmus algae. Scientific Reports. Nature Publishing Group, Vol 8(1), pp.10270. doi: 10.1038/s41598-018-28627-z.
Ciurli, A., Modeo, L., Pardossi, A. and Chiellini, C., (2021). Multidisciplinary integrated characterization of a native Chlorella-like microalgal strain isolated from a municipal landfill leachate. Algal Research, Vol 54, pp. 102202.
De Clerck, O. et al. (2013). Algal Taxonomy: A Road to Nowhere?’, Journal of Phycology. J Phycol, pp. 215–225. doi: 10.1111/jpy.12020.
Coleman, A. W. (2003). ITS2 is a double-edged tool for eukaryote evolutionary comparisons.
Coleman, A. W. (2009). Is there a molecular key to the level of “biological species” in eukaryotes? A DNA guide. Molecular Phylogenetics and Evolution. Academic Press, pp. 197–203. doi: 10.1016/j.ympev.2008.10.008.
de-Bashan, L. E. et al. (2008). Chlorella sorokiniana UTEX 2805, a heat and intense, sunlight-tolerant microalga with potential for removing ammonium from wastewater. Bioresource Technology, Vol 99(11), pp. 4980–4989. doi: 10.1016/j.biortech.2007.09.065.
Dimitrova, P. et al. (2017). Biochemical Characteristics of a Newly Isolated Strain Coelastrella Sp. Bgv Cultivated At Different Temperatures and Light Intensities. Vol 102(4), pp. 139–146.
Dörr, R. and Huss, V. A. R. (1990). Characterization of nuclear DNA in 12 species of Chlorella (Chlorococcales, Chlorophyta) by DNA reassociation. BioSystems, Vol 24(2), pp. 145–155. doi: 10.1016/0303-2647(90)90007-N.
Draaisma, R. B. et al. (2013). Food commodities from microalgae. Current Opinion in Biotechnology, pp. 169–177. doi: 10.1016/j.copbio.2012.09.012.
Elliott, L. G. et al. (2012). Establishment of a bioenergy-focused microalgal culture collection. Algal Research, Vol 1(2), pp. 102–113. doi: 10.1016/j.algal.2012.05.002.
Enamala, M. K. et al. (2018). Production of biofuels from microalgae – A review on cultivation, harvesting, lipid extraction, and numerous applications of microalgae. Renewable and Sustainable Energy Reviews, Vol 94 pp. 49–68. doi: 10.1016/j.rser.2018.05.012.
Feng, P. et al. (2014). Characterization of Chlorococcum pamirum as a potential biodiesel feedstock. Bioresource Technology, Vol 162. doi: 10.1016/j.biortech.2014.03.076.
Furnas, M. J. (1990). In situ growth rates of marine phytoplankton: approaches to measurement, community and species growth rates. Journal of Plankton Research, Vol 12(6), pp. 1117–1151. doi: 10.1093/plankt/12.6.1117.
Gill, S. S. et al. (2016). Strain selection, growth productivity and biomass characterization of novel microalgae isolated from fresh and wastewaters of upper Punjab, Pakistan’. Frontiers in Life Science, Vol 9(3), pp. 190–200. doi: 10.1080/21553769.2016.1204957.
Goecke, F. et al. (2020). Revision of Coelastrella (Scenedesmaceae, Chlorophyta) and first register of this green coccoid microalga for continental Norway. World Journal of Microbiology and Biotechnology, Vol 36(10), pp. 1–17. doi: 10.1007/s11274-020-02897-0.
Gonzalez, M. A. et al. (2001). Phylogenetic relationship among various strains of Dunaliella (Chlorophyceae) based on nuclear its rDNA sequences. Journal of Phycology Vol 37(4), pp. 604–611. doi: 10.1046/j.1529-8817.2001.037004604.x.
Gour, R. S. et al. (2016). Characterization and screening of native Scenedesmus sp. isolates suitable for biofuel feedstock. PLoS ONE, Vol 11(5), pp. 1–16. doi: 10.1371/journal.pone.0155321.
Guerin, M., Huntley, M. E. and Olaizola, M. (2003). Haematococcus astaxanthin: Applications for human health and nutrition. Trends in Biotechnology, pp. 210–216. doi: 10.1016/S0167-7799(03)00078-7.
Guillard, R. R. and Ryther, J. H. (1962). Studies of marine planktonic diatoms. I. Cyclotella nana Hustedt, and Detonula confervacea (cleve) Gran. Canadian journal of microbiology, Vol 8,cpp. 229–239. doi: 10.1139/m62-029.
Hanagata, N. (1998). Phylogeny of the subfamily Scotiellocystoideae (Chlorophyceae, Chlorophyta) and related taxa inferred from 18s ribosomal RNA gene sequence data. Journal of Phycology, Vol 34(6), pp. 1049–1054. doi: 10.1046/j.1529-8817.1998.341049.x.
Van Hannen, E. J., Fink, P. and Lürling, M. (2002). A revised secondary structure model for the internal transcribed spacer 2 of the green algae Scenedesmus and Desmodesmus and its implication for the phylogeny of these algae. European Journal of Phycology, Vol 37(2), pp. 203–208. doi: 10.1017/S096702620200361X.
Hegewald, E. et al. (2005). Revision of the Desmodesmus (Sphaeropleales, Scenedesmaceae) species with lateral spines. 2. The multi-spined to spineless taxa. Algological Studies/Archiv für Hydrobiologie, Supplement Vol 116, pp. 1–38. doi: 10.1127/1864-1318/2005/0116-0001.
Hegewald, E. et al. (2010). ITS2 sequence-structure phylogeny in the Scenedesmaceae with special reference to Coelastrum (Chlorophyta, Chlorophyceae), including the new genera Comasiella and Pectinodesmus. Phycologia, Vol 49(4), pp. 325–335. doi: 10.2216/09-61.1.
Hegewald, E., Bock, C. and Krienitz, L. (2013). A phylogenetic study on Scenedesmaceae with the description of a new species of Pectinodesmus and the new genera Verrucodesmus and Chodatodesmus (Chlorophyta, Chlorophyceae). Fottea. Czech phycological Society, Vol 13(2), pp. 149–164. doi: 10.5507/fot.2013.013.
Hoshina, R. (2014). DNA analyses of a private collection of microbial green algae contribute to a better understanding of microbial diversity. BMC Research Notes, VOL 7(1), pp. 1–15. doi: 10.1186/1756-0500-7-592.
Hoshina, R., Iwataki, M. and Imamura, N. (2010). Chlorella variabilis and Micractinium reisseri sp. nov. (Chlorellaceae, Trebouxiophyceae): Redescription of the endosymbiotic green algae of Paramecium bursaria (Peniculia, Oligohymenophorea) in the 120th year. Phycological Research. Blackwell Publishing, Vol 58(3), pp. 188–201. doi: 10.1111/j.1440-1835.2010.00579.x.
Hosseini Tafreshi, A. and Shariati, M. (2009). Dunaliella biotechnology: Methods and applications. Journal of Applied Microbiology, pp. 14–35. doi: 10.1111/j.1365-2672.2009.04153.x.
Hu, C. W. et al. (2013a). Pigment production by a new thermotolerant microalga Coelastrella sp. F50. Food Chemistry Vol 138(4), pp. 2071–2078. doi: 10.1016/j.foodchem.2012.11.133.
Hu, C. W. et al. (2013b). Pigment production by a new thermotolerant microalga Coelastrella sp. F50. Food Chemistry Vol 138(4), pp. 2071–2078. doi: 10.1016/j.foodchem.2012.11.133.
Jeon, S. L. and Hegewald, E. (2006). A revision of the species Desmodesmus perfaratus and D. tropicus (Scenedesmaceae, Chlorophyceae, Chlorophyta). Phycologia, Vol 45(5), pp. 567–584. doi: 10.2216/05-63.1.
Jiang, L., Li, Y. and Pei, H., (2021). Algal–bacterial consortia for bioproduct generation and wastewater treatment. Renewable and Sustainable Energy Reviews, Vol 149, p.111395.
Johnson, M. K. et al. (1968). Effects of Salts on the Halophilic Alga Dunaliella viridis. Journal of Bacteriology, Vol 95 (4), pp. 1461-1468. doi: 10.1128/JB.95.4.1461-1468.1968
Kalina, T. and Punčochářová (1987). Taxonomy of the subfamily Scotiellocystoideae Fott 1976 (Chlorellaceae, Chlorophyceae). Algological Studies, pp. 473–521.
Kaspar, H. F. et al. (2014). Continuous production of Chaetoceros calcitrans in a system suitable for commercial hatcheries. Aquaculture Vol 420 (421), pp. 1–9.doi: 10.1016/j.aquaculture.2013.10.021.
Karm, L.I.F.A. and Dwaish, A.S., (2021). Investigation of Some Ecological Factors and Isolation Techniques for Some Local Algae in Iraq. Annals of the Romanian Society for Cell Biology, pp.1059-1068.
Kaufnerová, V. and Eliáš, M. (2013). The demise of the genus Scotiellopsis Vinatzer (Chlorophyta). Nova Hedwigia, Vol 97(3–4), pp. 415–428. doi: 10.1127/0029-5035/2013/0116.
Kawasaki, S. et al. (2013). A Novel Astaxanthin-Binding Photooxidative Stress-Inducible Aqueous Carotenoprotein from a Eukaryotic Microalga Isolated from Asphalt in Midsummer. Plant and Cell Physiology. Oxford Academic, Vol 54(7), pp. 1027–1040. doi: 10.1093/pcp/pct080.
Kawasaki, S. et al. (2020). Coelastrella astaxanthina sp. nov. (Sphaeropleales, Chlorophyceae), a novel microalga isolated from an asphalt surface in midsummer in Japan’, Phycological Research. Blackwell Publishing, Vol 68(2), pp. 107–114. doi: 10.1111/pre.12412.
Kessler, E. (1985). Upper limits of temperature for growth in Chlorella (Chlorophyceae). Plant Systematics and Evolution. Springer-Verlag, Vol 151(1–2), pp. 67–71. doi: 10.1007/BF02418020.
Kessler, E. and Huss, V. A. R. (1992). Comparative physiology and biochemistry and taxonomic assignment of the Chlorella (Chlorophyceae) strains of the culture collection of the University of Texas at Austin. Journal of Phycology, Vol 28(4), pp. 550–553. doi: 10.1111/j.0022-3646.1992.00550.x.
Khan, S. A. et al. (2009). Prospects of biodiesel production from microalgae in India. Renewable and Sustainable Energy Reviews, Vol 13(9), pp. 2361–2372. doi: 10.1016/j.rser.2009.04.005.
Khatoon, H. et al. (2020). Growth and carotenoid production of Dunaliella salina (Dunal) teodoresco, 1905 cultured at different salinities. Asian Fisheries Science, Vol 33(3), pp. 207–212. doi: 10.33997/j.afs.2020.33.3.001.
Knothe, G. (2010). Biodiesel and renewable diesel: A comparison. Progress in Energy and Combustion Science, Vol 36(3), pp. 364–373. doi: 10.1016/j.pecs.2009.11.004.
Krienitz, L. and Bock, C. (2012). Present state of the systematics of planktonic coccoid green algae of inland waters. Hydrobiologia. pp. 295–326. doi: 10.1007/s10750-012-1079-z.
Krienitz, L., Huss, V. A. R. and Bock, C. (2015). Chlorella: 125 years of the green survivalist. Trends in Plant Science, pp. 67–69. doi: 10.1016/j.tplants.2014.11.005.
Kunz, W. F. (1972). Response of the alga Chlorella sorokiniana to 60Co gamma radiation. Nature, Vol 236(5343), pp. 178–179. doi: 10.1038/236178a0.
Lee, H. G. et al. (2016). Unique mitochondrial genome structure of the green algal strain yc001 (Sphaeropleales, Chlorophyta), with morphological observations. Phycologia, Vol 55(1), pp. 72–78. doi: 10.2216/15-71.1.
Lemieux, C., Otis, C. and Turmel, M. (2014). Chloroplast phylogenomic analysis resolves deep-level relationships within the green algal class Trebouxiophyceae. BMC Evolutionary Biology, Vol 14(1), p. 211. doi: 10.1186/s12862-014-0211-2.
Lizzul, A. M. et al. (2014). Combined remediation and lipid production using Chlorella sorokiniana grown on wastewater and exhaust gases. Bioresource Technology. Vol 151, pp. 12–18. doi: 10.1016/j.biortech.2013.10.040.
Lizzul, A. M. et al. (2018). Characterization of Chlorella sorokiniana, UTEX 1230. Biology, Vol 7(2), pp. 1–12. doi: 10.3390/biology7020025.
Luo, L. et al. (2016). Nutrient removal and lipid production by Coelastrella sp. in anaerobically and aerobically treated swine wastewater. Bioresource Technology, Vol 216, pp. 135–141. doi: 10.1016/j.biortech.2016.05.059.
Luo, W. et al. (2010). Generic concept in Chlorella-related coccoid green algae (Chlorophyta, Trebouxiophyceae). Plant Biology, Vol 12(3), pp. 545–553. doi: 10.1111/j.1438-8677.2009.00221.x.
Markou, G. and Nerantzis, E. (2013). Microalgae for high-value compounds and biofuels production: A review with focus on cultivation under stress conditions. Biotechnology Advances, pp. 1532–1542. doi: 10.1016/j.biotechadv.2013.07.011.
Mata, T. M., Martins, A. A. and Caetano, N. S. (2010). Microalgae for biodiesel production and other applications: A review’, Renewable and Sustainable Energy Reviews, 14(1), pp. 217–232. doi: 10.1016/j.rser.2009.07.020.
Melkonian, M. and Preisig, H. R. (1984). Ultrastructure of the flagellar apparatus in the green flagellate Spermatozopsis similis. Plant Systematics and Evolution, Vol 146(3–4), pp. 145–162. doi: 10.1007/BF00989542.
Mulders, K. J. M. et al. (2014). Phototrophic pigment production with microalgae: Biological constraints and opportunities. Journal of Phycology, Vol 50(2), pp. 229–242. doi: 10.1111/jpy.12173.
Neofotis, P. et al. (2016). Characterization and classification of highly productive microalgae strains discovered for biofuel and bioproduct generation. Algal Research, Vol 15, pp. 164–178. doi: 10.1016/j.algal.2016.01.007.
Nozaki, H. et al. (1995). Taxonomic re-examination of the two strains labeled “Chlorella” in the Microbial Culture Collection at the National Institute for Environmental Studies (NIES-Collection). Microbiol. Cult. Coll, Vol 11(1), pp. 11–18.
Olaizola, M. (2003). Commercial development of microalgal biotechnology: From the test tube to the marketplace. In Biomolecular Engineering, pp. 459–466. doi: 10.1016/S1389-0344(03)00076-5.
Oren, A. (2014). The ecology of Dunaliella in high-salt environments. Journal of Biological Research (Greece). doi: 10.1186/s40709-014-0023-y.
Park, K. C. et al. (2012). Mixotrophic and photoautotrophic cultivation of 14 microalgae isolates from Saskatchewan, Canada: Potential applications for wastewater remediation for biofuel production. Journal of Applied Phycology, Vol 24(3), pp. 339–348. doi: 10.1007/s10811-011-9772-2.
Pertumbuhan, P. et al. (2017). Growth Evaluation of Microalgae Isolated From Palm Oil Mill Effluent in Synthetic Media. Malaysian Journal of Analytical Science, Vol 21(1), pp. 82–94. doi: 10.17576/mjas-2017-2101-10.
Polle, J. E. W. et al. (2017). Draft nuclear genome sequence of the halophilic and beta-carotene-accumulating green alga Dunaliella salina strain CCAP19/18. Genome Announcements. American Society for Microbiology. doi:10.1128/genomeA.01105-17.
Punčochářová, M. and Kalina, T. (1981). Taxonomy of the genus Scotiellopsis Vinatzer (Chlorococcales, Chlorophyta). Algological Studies, Vol 27, pp. 119–147. doi: 10.1127/algol_stud/27/1981/119.
Ratha, S. K. et al. (2012). Bioprospecting and indexing the microalgal diversity of different ecological habitats of India. World Journal of Microbiology and Biotechnology, Vol 28(4), pp. 1657–1667. doi: 10.1007/s11274-011-0973-2.
Ratha, S. K. and Prasanna, R. (2012). Bioprospecting microalgae as potential sources of “Green Energy”-challenges and perspectives (Review). Applied Biochemistry and Microbiology, pp. 109–125. doi: 10.1134/S000368381202010X.
Richmond, A. (2003). Handbook of microalgal cultures, Blackwell Publishers, Boston. doi: 10.1002/9780470995280.
Rippka, R., Deruelles, J. and Waterbury, J. B. (1979). Generic assignments, strain histories and properties of pure cultures of cyanobacteria. Journal of General Microbiology, Vol 111(1), pp. 1–61. doi: 10.1099/00221287-111-1-1.
Rodolfi, L. et al. (2009). Microalgae for oil: Strain selection, induction of lipid synthesis and outdoor mass cultivation in a low-cost photobioreactor. Biotechnology and Bioengineering, Vol 102(1), pp. 100–112. doi: 10.1002/bit.22033.
SAG Göttingen (2013). Kuhl-Medium für unizelluläre Grünalgen. pp. 1–3. Available at: http://sagdb.uni-goettingen.de/culture_media/12 Unicellular Green Algae Medium.pdf.
Sajjadi, B. et al. (2018). Microalgae lipid and biomass for biofuel production: A comprehensive review on lipid enhancement strategies and their effects on fatty acid composition. Renewable and Sustainable Energy Reviews, Vol 97 pp. 200–232. doi: 10.1016/j.rser.2018.07.050.
Schultz, J. et al. (2006). The internal transcribed spacer 2 database – A web server for (not only) low level phylogenetic analyses. Nucleic Acids Research, Vol 34 pp 704. doi: 10.1093/nar/gkl129.
Schultz, J. and Wolf, M. (2009). ITS2 sequence-structure analysis in phylogenetics: A how-to manual for molecular systematics. Molecular Phylogenetics and Evolution. Academic Press, pp. 520–523. doi: 10.1016/j.ympev.2009.01.008.
Serediak, N. and Huynh, M. (2011). Algae identification lab guide : accompanying manual to the algae identification field guide, Agriculture and Agri-Food Canada, Ottawa (Ontario).
Sime, I. (2004). The freshwater algal flora of the British Isles: An identification guide to freshwater and terrestrial algae, edited by David M. John, Brian A. Whitton and Alan J. Brook. Cambridge University Press, Cambridge, 2002, 702pp. ISBN 0-521-77051-3. Aquatic Conservation: Marine and Freshwater Ecosystems, Vol 14(1) pp. 105–105. doi: 10.1002/aqc.579.
Song, M. et al. (2013). Evaluation of the potential of 10 microalgal strains for biodiesel production. Bioresource Technology, Vol 141 pp. 245–251. doi:10.1016/j.biortech.2013.02.024.
Sorokin, C. and Myers, J. (1953). A high-temperature strain of Chlorella. American Association for the Advancement of Science, Vol 117(3039) pp. 330–331. doi: 10.1126/science.117.3039.330.
Thao, T. Y. et al. (2017). Isolation and selection of microalgal strains from natural water sources in Viet Nam with potential for edible oil production. Marine Drugs. MDPI AG, Vol 15(7). doi: 10.3390/md15070194.
Tschaikner, A. G. and Kofler, W. (2008). Coelastrella aeroterrestrica sp. nov. (Chlorophyta, Scenedesmoideae) a new, obviously often overlooked aeroterrestrial species. Algological Studies. Vol 128, pp. 11–20. doi: 10.1127/1864-1318/2008/0128-0011.
Wang, Q. et al. (2019). Morphology and molecular phylogeny of coccoid green algae Coelastrella sensu lato (Scenedesmaceae, Sphaeropeales), including the description of three new species and two new varieties. Journal of Phycology. Blackwell Publishing Inc, Vol 55(6), pp. 1290–1305. doi: 10.1111/jpy.12915.
Wijffels, R. H. and Barbosa, M. J. (2010). An outlook on microalgal biofuels. American Association for the Advancement of Science, pp. 796–799. doi:10.1126/science.1189003.
Wijffels, R. H., Kruse, O. and Hellingwerf, K. J. (2013). Potential of industrial biotechnology with cyanobacteria and eukaryotic microalgae. Current Opinion in Biotechnology, pp. 405–413. doi: 10.1016/j.copbio.2013.04.004.
Yin, Z. et al. (2020). A comprehensive review on cultivation and harvesting of microalgae for biodiesel production: Environmental pollution control and future directions. Bioresource Technology, Vol 301 p. 122804. doi: 10.1016/j.biortech.2020.122804.
Yuan, J. P. et al. (2011). Potential health-promoting effects of astaxanthin: A high-value carotenoid mostly from microalgae. Molecular Nutrition and Food Research. pp. 150–165. doi: 10.1002/mnfr.201000414.
Zachleder, V., Bisova, K. and Vitova, M. (2016). The Cell Cycle of Microalgae, The Physiology of Microalgae. pp 3-46. doi: 10.1007/978-3-319-24945-2.
Zhang, B. et al. (2014). Characterization of a native algae species Chlamydomonas debaryana: Strain selection, bioremediation ability, and lipid characterization. BioResources, Vol 9(4), pp. 6130–6140. doi: 10.15376/biores.9.4.6130-6140.


