Department of Biology, College of Science, University of Ha’il, Ha’il-2440, Kingdom of Saudi Arabia
Article Publishing History
Received: 08/04/2020
Accepted After Revision: 25/05/2020
Helicobacter pylori (H. pylori) bacterium is widely studied risk factor of gastric cancer. The mechanism by which H. pylori induces gastric carcinogenesis, has been studied by various researcher. However, a molecular level mechanism by which H. pylori enters into the cells causes p53 modulation leads to stress and gastric cancer developments, is still an open question. In the present studied we developed p53-ROS-H. pylori model integrated deterministic model to show the molecular level interaction between p53 proteins network and H. pylori bacterium via reactive oxygen species. The interactions among p53 network proteins, reactive oxygen species and H. pylori are described by system of a set of ordinary differential equations. The rate of the reaction has been set by using pervious experimental and theoretical works. These sets of differential equations further numerically solved by using standard Runge-Kutta fourth order method at high computational cost. Numerical simulation results showed that as the accumulation of H. pylori (kHP) in the cells increases, the instability of p53 network systems are also increase. Various phases of the system have been observed viz normal phase, stable to damp phase, damped to stable phase and stationary phase. These phases correspond to various stages of cell cycle. The present model clarifies the molecular level interactions between H. pylori via reactive oxygen species. This model suggests that as the accumulation of H. pylori reaches at a particular threshold value, it leads to the cancerous progression. The suggested model will be very useful to understand the temporal dynamics of gastric cancer progression due to H. pylori bacterium. Further, study is needed in realistic environment.
Helicobacter pylory, p53, Gastric Cancer, DNA damage, Temporal Dynamics
Alam M. J. Helicobacter Pylori Leads to the Activation of Host Cell Intrinsic P53 Network Via Dna Damage: A Deterministic Model. Biosc.Biotech.Res.Comm. 2020;13(2).
Alam M. J. Helicobacter Pylori Leads to the Activation of Host Cell Intrinsic P53 Network Via Dna Damage: A Deterministic Model. Biosc.Biotech.Res.Comm. 2020;13(2). Available from: https://bit.ly/31lzatB
Copyright © Alam This is an open access article distributed under the terms of the Creative Commons Attribution License (CC-BY) https://creativecommns.org/licenses/by/4.0/, which permits unrestricted use distribution and reproduction in any medium, provide the original author and source are credited.
INTRODUCTION
The expression of p53 tumor suppressor protein in the cell is reported as marker of the most of the cancer (Blagih et. al., 2020). It acts as a key regulator in the cellular network and response to a variety of cellular stress, including DNA damage, hypoxia, nucleotide depletion, nitric oxide and aberrant proliferative signals (such as oncogene activation) (Moll and Petrenko, 2003). But in most cancerous cells, p53 tumor suppressor signaling pathway usually found in inactivated condition (Blagih et. al., 2020). In normal cells, stress induced p53 leads to the participation in various key cellular processes, such as, cell-cycle arrest, senescence and most importantly tumor clearance to prevent cancer cell formation (Leeuwen, 2020, Zambetti, 2007). Moreover, it is reported that, activated p53 protein safeguards the organism against the propagation of cells that carry damaged DNA with potentially oncogenic mutations (Moll and Petrenko, 2003).
p53 act as transcription factor of MDM2 protein. Moreover, MDM2 act as negative feedback regulator of p53 protein which is also considered as an effective approach in cancer therapy (Leeuwen, 2020, Michael and Oren, 2003). MDM2 physically blocking its ability binding itself to p53 leads to the transactivation of p53 gene expression, and stimulating its degradation (Fang et. al., 2000). Further, the interaction of N-terminal domain of MDM2 with transactivation domain of p53 (p53TAD) performs a significant role in the regulation of the G1 checkpoint of the cell cycle and cell function (Boyd et. al., 2000). ROS (Reactive oxygen species) are chemically reactive molecules containing oxygen ions and peroxides (Amendola et. al., 2013). They are synthesized from normal metabolism of oxygen as a natural byproduct and play important roles in cell signaling and homeostasis (Devasagayam et. al., 2004; Rowe et. al., 2008 ans Poetsh, 2020).
However, ROS level inside cell can be elevated by UV irradiation or heat exposure which can drive the cell at different stress states (Devasagayam et. al., 2004) High level of ROS can promote DNA damage, and may probably lead the cell to mutagenesis, carcinogenesis and aging (Poetsh, 2020, Amendola et. al., 2013; Rowe et. al., 2008; Proctor and Gray, 2010). H. pylori is a structurally spiral shaped bacterium. It generally present in digestive tract of the human. It is reported that in developing country children are commonly infected by H. pylori (Horvat et.al., 2018; Kakelar et.al., 2019, Choi, et. al, 2020).
Studies suggests that control of H. pylori leads to the control of gastric cancer (Ford et. al., 2020, Hu et. al., 2020; Sablet et.al., 2011). The inclusion of H. pylori in the host cells increases the production of ROS, which further leads to the suppression of the endogenous level of p53 protein via DNA damage and represses to apoptosis in human gastric cells (Handa et. al. 2011; Polk and Peek, 2010). Various studies have been performed to understand the mechanism by which H. pylori induces gastric carcinogenesis (Wang et. al., 2020, Handa et. al., 2011, Achanta et. al., 2004). However, how the dynamical behaviour of p53 protein changes at molecular level due to H. pylori via reactive oxygen species, is still an open question. In the present study, we try to answer these fundamental questions based on basic deterministic model.
MATERIAL AND METHOD
p53-MDM2-H. pylori model.- The model we consider (Fig. 1) is integration of p53-Mdm2 regulatory network (Proctor and Gray, 2008) with stress inducers ROS via DNA damage (Rowe et. al., 2008) and H. pylori (Handa et. al., 2011). In this model we assume that H. pylori are supposed to be constantly entered in the nucleus after infection. ROS synthesis due to H. pylori is assumed to occur with a rate of k12. This ROS synthesis triggers DNA damage with a rate of k14 (Rowe et. al., 2008). Then this DNA damage leads to the activation of ARF with a rate k16 (Lee et. al., 2005) followed by the degradation of ARF with a rate of k17. Further, the activated ARF protein binds to MDM2 with a rate of k18 to control ubiquitination of p53 (Zhang and Xiong, 2001) The ARF and MDM2 interaction results into to the formation of ARF_MDM2 complex (Khan et. al., 2004).
The formation of ARF_MDM2 complex reduces the concentration level of MDM2 in the system which in turn alters the behaviour of p53 (Khan et. al., 2004). On the other hand, dissociation of ARF_MDM2 complex with a rate k19 helps the degradation of MDM2 population and recruit activated ARF. The synthesis of p53 takes place through transcription of p53_mRNA with a rate k2. Further, this p53 synthesis depends on the available p53_mRNA concentration level. At normal condition p53 is generally bound to MDM2 with a rate k8 recruiting a complex p53_MDM2 and after which the dissociation of the complex ubiquitinates p53 with a rate k9 and MDM2 with a rate k12 (Zatorsky et. al., 2006) exhibiting oscillatory behavior of p53 in the model. Further, rate of MDM2 transcription by p53 via production of MDM2_mRNA, at k5 (Proctor and Gray, 2008).
Hence, the MDM2_mRNA provides intermediary link between p53 and MDM2. The self ubiquitination of MDM2_mRNA is assumed to be with a rate k7. MDM2_mRNA synthesize MDM2 protein with a rate k6. The self ubiquitination of MDM2 is assumed to be with a rate k11. The molecular species involved in this model are listed in Table 1, and the biochemical reaction channels involved in the model network with their descriptions, kinetic laws and values of the rate constants used in our simulations are given in Table 2.The model biochemical network (Fig. 1) described by the twenty two reaction channels (Table 2) can be described by the following coupled ordinary differential equations (ODE) using Mass action law of chemical kinetics,
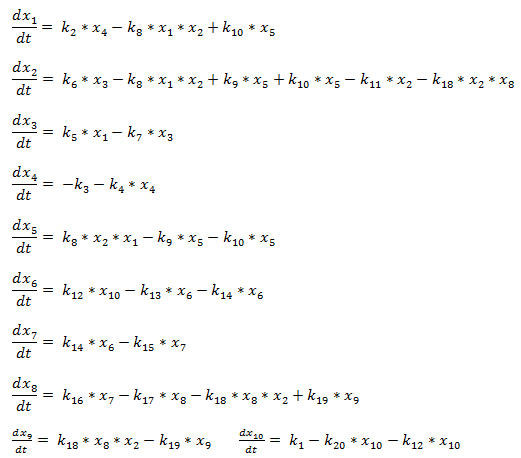
The set of ODEs can be written in compact form as in the following,
![]() where
where ![]() is the functional vector.
is the functional vector.
The time evolution of the state vector ![]() can be obtained by numerically solving the non-linear coupled differential equations (1)-(11) using standard 4th order Runge-Kutta algorithm for numerical integration (Press, 1992).Consider the state of the system be described by a state vector given by,
can be obtained by numerically solving the non-linear coupled differential equations (1)-(11) using standard 4th order Runge-Kutta algorithm for numerical integration (Press, 1992).Consider the state of the system be described by a state vector given by,
![]() where, {x} is the set of concentrations of the respective molecular species, N=10 and T is the transpose of the vector. The model biochemical network (Fig. 1) described by the twenty reaction channels (Table 2) can be described by the following coupled ordinary differential equations (ODE) using Mass action law of chemical kinetics,
where, {x} is the set of concentrations of the respective molecular species, N=10 and T is the transpose of the vector. The model biochemical network (Fig. 1) described by the twenty reaction channels (Table 2) can be described by the following coupled ordinary differential equations (ODE) using Mass action law of chemical kinetics,
![]()
where, i=1,2,…,N and Fi is the ith function whose form is given in table. The non-linear coupled N ODEs of p53-MDM2-H. pylori model are solved using 4th order Runge-Kutta method which is the standard algorithm for numerical integration (Press, 1992) to find the dynamics of the system variables. The simulation is done for 10 days using the parameter values given in Table 2 and starting from an initial condition.
Table 1. List of molecular species
| S.No. | Species Name | Description | Notation |
| 1. | p53 | Unbounded p53 protein | x1 |
| 2. | MDM2 | Unbounded Mdm2 protein | x2 |
| 3. | MDM2_mRNA | Mdm2 messenger mRNA | x3 |
| 4. | p53_mRNA | p53 messenger mRNA | x4 |
| 5. | p53_MDM2 | Mdm2 with p53 complex | x5 |
| 6. | ROS | Reactive Oxygen Species | x6 |
| 7. | Dam_DNA | Damage DNA | x7 |
| 8. | ARF | Alternative Reading Frame protein | x8 |
| 9. | ARF_MDM2 | ARF and Mdm2 complex | x9 |
| 10. | HP | Helicobacter pylory facror | x10 |
Table 2. List of chemical reaction, Kinetic Laws and their rate constant
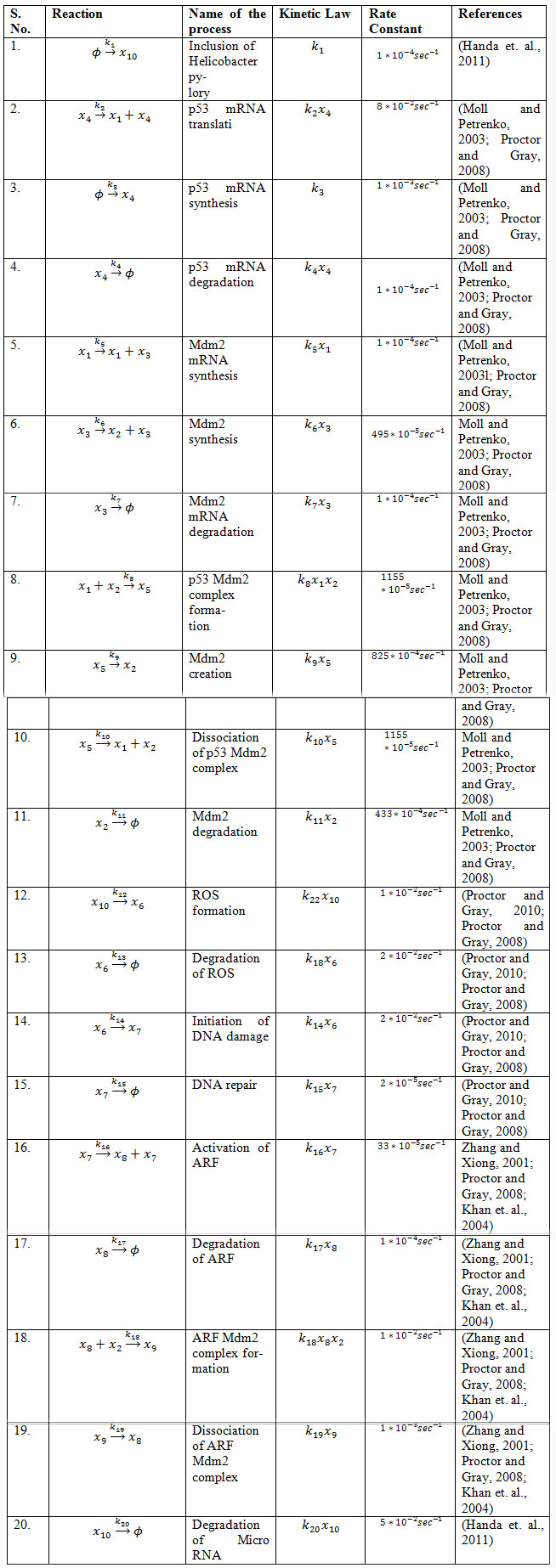
Figure 1: A schematic diagram of p53-Mdm2-H. pylori network model.
RESULTS AND DISCUSSION
Role of H. pylori on p53 dynamics: The impact of H. pylori on p53 was studied by keeping fixed k6=0.0001 and allowing to change the values of k10 (Fig. 2). We got three different states namely stable, damped with sustain oscillation and again stable state of p53 driven by H. pylori (Fig. 2). The small values of k10 (K10<0>) could not able to provide significant stress to p53 dynamics, and maintains at stabilized state (Fig. 2 E). The further increase in k10 values (<0.005>k10<0.1>) the dynamics still maintains stability upto certain interval of time (Fig. 2 B, C, D), after which the dynamics is switched to damped oscillation (weakly activated) for short interval of time and then to sustain oscillation (strongly activated). Further increase in k10 compels the dynamics to stabilized state again with low concentration level (Fig. 2 A).
This suggests that the increase in concentration of H. pylori (k10 coresponds to kHP) in the system drives the system at various stress states, lowering p53 concentration level (Wei et. al., 2010; Polk and Peek, 2010; Handa et. al., 2011). Moreover suggests that extreme values of k_ROS may cause very high DNA damage, such that the damage could not able be repaired back, which could be the condition of apoptotic phase. The excess kHP values induce lowering of p53 concentration level even below normal stabilized p53 state indicating the possibility of switching stress state to cancerous state (Handa et. al., 2011).
Figure 2: The p53 temporal behaviour when H. pylori act as a stress inducer.
Modulation of p53 dynamics by H. pylori: The impact of H. pylori on the amplitude and time period of p53 dynamics is studied (Fig. 3) in order to understand modulation of p53 dynamics driven by stress inducer. The impact of k_HP on amplitude and time period of p53 dynamics is shown (Fig. 3). The lower values in k_HP could not able to make significant change in amplitude of p53 dynamics, and maintains at stable state with high value of constant p53 amplitude (Fig. 3). Further increase in k_HP drives the p53 amplitude to decrease monotonically, and then reach a stable state with minimum amplitude. The behaviour of the time period T_p53 as a function of k_HP shows three regimes, stable, monotonically decreased with increase in k_HP and stable state again. However the different behaviour i.e. the decrease in A_p53 as increase in k_HP may trigger cancer state from stress condition.
Figure 3: A comparative plot for the amplitude verses k_HP and the time period variation verses k_HP.
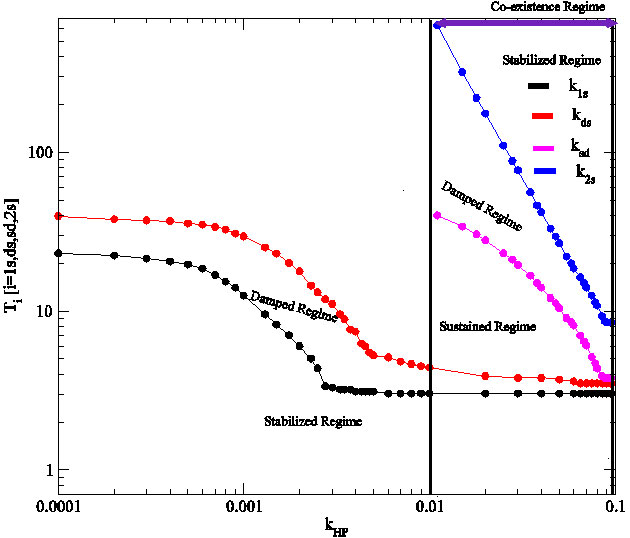
Co-existence of states: The phase transition like behaviour of the system dynamics induced by H. pylori concentrations available in the system can be well characterized by analyzing the nature of transition time of the p53 dynamics. We define T_1s to be the transition time below () which the dynamics shows stable state (does not show any oscillation) and above which the dynamics shows oscillatory behaviour. We further define second transition time, T_ds which separates increasing damped and sustain oscillations (Fig. 2). Similarly, T_sd and T_2s are taken as transition times separating sustain and damped oscillation, and damped oscillation and stabilized state. We then calculated T_1s, T_ds, T_sd and T_2s as a function of kHP (Fig. 4 upper panel) where the regimes for T< T_{1s} > and T < T_{2s} > corresponds to stabilized states, regimes between T_{ds} < T < T_{1s} and T_{2s} < T < T_{sd} corresponds to damped states and T_{sd}< T < T_{ds} indicates the sustain oscillation state regime.
Figure 4: A phase diagram showing impact of k_HP on stability of p53 protein.
The co-existence of the four states can be obtained in the case of H. pylori induced p53 dynamics. Within this co-existence regime, the regions of damped, sustain and stabilized states are observed (Fig. 4). The results indicate that there is a certain range of kHP (region bounded by two lines) where one can find the four states together including two stable states for any value of kHP (Fig. 4). This means that for any concentration of ROS which is due to H. pylori in the system corresponding to any values within this range, the p53 dynamics will stay stable for some interval of time, then it will start activated to reach maximum activation within certain interval of time and after sometime it will stay stable again. In the other regimes, at most we can find three states. This co-existence of the states indicate that exposure of the system to constant H. pylori concentration can drive the system from normal to stress and then to apoptosis.
CONCLUSION
The variation in concentration of reactive oxygen species in cellular system triggers to the changes in the p53 dynamics (various stress states). Further, the introduction H. pylori to the system shows inhibitory effect on p53 production and switching of stress states by varying H. pylori concentration. Present model suggests that H. pylori accumulation in host gastric cells, triggers cancerous progression. The obtained results are quite interesting and provide hidden molecular level information regarding the activity of H. pylori that it can probably switch the system to cancerous state. Moreover, the impact of the H. pylori on p53 regulatory pathway should be further studied in realistic system in order to capture the state switching mechanism quantitatively and to understand the role of noise in the cellular process.
ACKNOWLEDGMENTS
This research has been funded by Scientific Research Deanship at University of Ha’il, Saudi Arabia, through research project number: 160667.
REFERENCES
Achanta G, and Huang P. (2004). Role of p53 in Sensing Oxidative DNA Damage in Response to Reactive Oxygen Species-Generating Agents. Cancer Research; 64(17), 6233-6239.
Amendola R., Cervelli M., Fratini E., Sallustio D.E., Tempera G., Ueshima T., Mariottini P., and Agostinelli E. (2013). Reactive Oxygen Species Spermine Metabolites Generated From Amine Oxidases and Radiation Represent a Therapeutic Gain in Cancer Treatments. Int. J. Oncol.; 43(3), 813-820.
Blagih J., Buck M.D. and Vousden K.H. (2020). p53, cancer and the immune response Journal of Cell Science. J. Cell. Sc.; 133, 1-13.
Boyd M.T., Vlatkovic N., and Haines D.S. (2000). A Novel Cellular Protein (MTBP) Binds to MDM2 and Induces a G1 Arrest That Is Suppressed by MDM2. J. Biol. Chem.; 275(41), 31883-31890.
Choi I.J, Kim C.G., and Lee J.Y., Kim Y.I., Kook M.C., Park B., and Joo J. (2020). Family History of Gastric Cancer and Helicobacter pylori Treatment. N Engl J Med; 382, 427-436.
Devasagayam T.P., Tilak J.C., Boloor K.K., Sane K.S., Ghaskadbi S.S., and Lele R.D. (2004). Free radicals and antioxidants in human health: current status and future prospects. J. Assoc. Physicians India; 52, 794-804.
Fang S., Jensen J.P., Ludwig R.L., Vousden K.H., and Weissman A.M. (2000). Mdm2 Is a RING Finger-Dependent Ubiquitin Protein Ligase for Itself and p53. J. Biol. Chem.; 275(12), 8945-8951.
Ford A.C., Yuan Y., and Moayyedi P, (2020). Helicobacter pylori eradication therapy to prevent gastric cancer: systematic review and meta-analysis; Gut, 32089, 1-9.
Hu Y, Zhu Y, and Lu NH. (2020). Recent progress in Helicobacter pylori treatment. Chin Med J; 133 (3), 335–343.
Horvat A., Noto J. M., Ramatchandirin B., Zaika E., Palrasu M., Wei J, Schneider B. G., El-Rifai W., Peek Jr R. M., Zaika A. I. (2018). Helicobacter pylori pathogen regulates p14ARF tumor suppressor and autophagy in gastric epithelial cells. Oncogene; 37, 5054-5065.
Handa O., Naito Y., and Yoshikawa T. (2011). Redox Biology and Gastric Carcinogenesis: The Role of Helicobacter Pylori. Redox Rep., 16(1), 1-7.
Kakelar H.M., Barzegari A., Dehghani J., Hanifian S., Saeedi N., Barar J., Omidi Y. (2019). Pathogenicity of Helicobacter pylori in cancer development and impacts of vaccination. Gastric Cancer; 22, 23-36.
Khan S., Guevara C., Fujii G., and Parry D. (2004). p14ARF Is a Component of the p53 Response Following Ionizing Irradiation of Normal Human Fibroblasts. Oncogene, 23(36), 6040-6046.
Lee C., Smith B.A., Bandyopadhyay K., and Gjerset R.A. (2005). DNA Damage Disrupts the p14ARF-B23(nucleophosmin) Interaction and Triggers a Transient Subnuclear Redistribution of p14ARF. Cancer Res; 65(21), 9834-9842.
Leeuwen F.N.V. (2020). Therapeutic targeting of mutated p53 in acute lymphoblastic leukemia. Haematologica; 105(1), 10-11.
Michael D. and Oren M. (2003). The p53-MDm2 Module and the Ubiquitin System. Semin. Cancer Biol.; 13(1), 49-58.
Moll U.M. and Petrenko O. (2003). The MDM2-p53 Interaction. Mol. Cancer Res.; 1(14), 1001-1008.
Poetsch A.R. (2020). The genomics of oxidative DNA damage, repair, and resulting mutagenesis. Computational and Structural Biotechnology Journal; 18, 207–219.
Polk D.B., and Peek R. M. (2010). Helicobacter Pylori: Gastric Cancer and Beyond. Nat Rev Cancer; 10(6), 403-414.
Press W.H., Teukolsky S.A., Vetterling W.T, and Flannery B.P. (1992). Numerical Recipe in Fortran. Cambridge University Press.
Proctor C.J and Gray D.A. (2008). Explaining Oscillations and Variability in the p53-Mdm2 System. BMC Syst. Biol; 2(75), 1-20.
Proctor C.J. and Gray D.A. (2010) GSK3 and p53 – Is There a Link in Alzheimer’s Disease?, Molecular Neurodegeneration; 5(7) 1-15.
Rowe L.A., Degtyareva N., and Doetsch P.W. (2008). DNA Damage-Induced Reactive Oxygen Species (ROS) Stress Response in Saccharomyces Cerevisiae. Free Radic. Biol. Med.; 45(8), 1167-1177.
Sablet T., Piazuelo M.B., Shaffer C. L., Schneider B.G., Asim M., Chaturvedi . et al. (2011). Phylogeographic origin of Helicobacter pylori is a determinant of gastric cancer risk. Gut; 60(9), 1189-1195.
Smout D.T., Elliott T.B., Verspaget H.B., Jones D., Allen C.R., Vernon K.G., Bremner T., Kidd L.C.R, Kim K.S., Groupman J.D. and Ashktorab H. (2000). Influence of Helicobacter Pylori on Reactive Oxygen-Induced Gastric Epithelial Cell Injury. Carcinigenesis; 21(11), 2091-2095.
Wang S., Chenb Z., Zhub S., Lub H., Pengb D., Souttob M., Naz H., Peek R., Xua H., Zaikab A., Xua Z., El-Rifaib W. (2020). PRDX2 protects against oxidative stress induced by H. pylori and promotes resistance to cisplatin in gastric cancer. Redox biology, 28, 101319.
Wei J., Nagy T.A., Vilgelm A., Zaika E., Ogden S.R., Gallo J.R., Piazuelo M.N., Correa P., Washington M.K., El-Rifai W., Peek R.M., Zaika A. (2010). Regulation of p53 Tumor Suppressor by Helicobacter Pylori in Gastric Epithelial Cells. Gastroenterology; 139(4), 1333-1343.
Zambetti G.P. (2007). The p53 Mutation Gradient Effect and its Clinical Implication. J. Cell Physiol.; 213(2), 370-373.
Zatorsky N.G, Rosenfeld N., Itzkovitz S., Milo R., Sigal A., Dekel E., Yarnitzky T., Liron Y., Polak P., Lahav G., and Alon U. (2006). Oscillations and Variability in the p53 System. Mol Syst. Biol.; 2(2006.0033), 1-13.
Zhang Y. and Xiong Y. (2001). Control of p53 Ubiquitination and Nuclear Export by MDM2 and ARF. Cell Growth Differ; 12(4), 175-186.