Department of Biotechnology, Mizoram University, Aizawl, INDIA – 796004
Corresponding author email: mzut261@mzu.edu.in
Article Publishing History
Received: 15/04/2020
Accepted After Revision: 30/05/2020
The primary objective of this work has been to evaluate the expression level of recombinant protein in E.coli in the presence of different fusion tags. One of the major challenges in using bacterial host in overexpression of recombinant protein is the formation of inclusion bodies, therefore, resulting in biologically inactive protein. Tedious procedures of denaturation and refolding of inclusion bodies are required to obtain functional protein. In this work, a human gene is fused with different fusion tags such as nusA, endoxylanase signal, pelB leader and asparginase signal sequence and expressed in E.coli BL21(DE3) host cells. Results showed that nusA fusion showed highest expression level of total protein (approx. 80 mg g-1 DCW) while endoxylanase signal sequence demonstrated a high expression level of soluble protein (~40% of total protein). In the case of pelB leader fusion, the overall productivity was low with the insoluble fraction comprising the majority of the total target protein and ~ 16 % in soluble fraction. Also, in the case of asparaginase signal fusion, major part of the expressed target protein was in the insoluble fraction with approximately 12 % in soluble form. These results demonstrated that recombinant production in the presence of fusion tags leads to variation in the level of soluble and insoluble forms. There seemed to be a stringent regulation for the expression of soluble protein suggesting that the host machinery may favor the inclusion body formation possibly due to the toxicity of the recombinant product. This study would be helpful in optimization of genetic parameters for the selection of most suitable vector-host combinations, as well as further understanding of bacterial strategies in adaptation and survival to stress.
Recombinant, Fusion tags, Inclusion Bodies, E.coli
Vaiphei S. T. Evaluation of Fusion Tags for Recombinant Protein Expression in Bacterial System. Biosc.Biotech.Res.Comm. 2020;13(2).
Vaiphei S. T. Evaluation of Fusion Tags for Recombinant Protein Expression in Bacterial System. Biosc.Biotech.Res.Comm. 2020;13(2). Available from: https://bit.ly/3hbbBsG
Copyright © Vaiphei et al., This is an open access article distributed under the terms of the Creative Commons Attribution License (CC-BY) https://creativecommns.org/licenses/by/4.0/, which permits unrestricted use distribution and reproduction in any medium, provide the original author and source are credited.
INTRODUCTION
Approximately 70% of recombinant proteins are produced as inclusion bodies in E. coli over-expression system (Yang et al., 2011). In spite of having many advantages, inclusion body formation offers serious disadvantages such as the need for strong denaturants for solubilization and a subsequent refolding process to regain protein activity (Makrides, 1996; Singh et al., 2015, 2020). Moreover, refolded protein may not regain its biological activity and often resulted in reduction of protein yield even under optimized conditions of buffer composition, protein concentration, temperature, pH or ionic strength (Makrides, 1996, Lilie et al., 1998). It has been therefore, desirable to over-express recombinant protein in its soluble or active form, and thereby avoid the trial-and-error procedures required to develop an efficient refolding process (Kim et al., 2005). However, optimized solubilization method of inclusion bodies has also been reported for the retention of native secondary structure (Nekoufar et al., 2020).
Interferon-g (IFN-g) also known as a Type II interferon is secreted by lymphocytes against mitogenic stimulation and is involved in differentiation, proliferation and maturation of hematopoeitic cells. It also enhances non-specific immunity to tumors, as well as to microbial, viral and parasitic infections (Mamame et al., 1999, Sen and Lengyel, 1992). There have been several reports regarding the production of recombinant human interferon-gamma (rhIFN-g) in E.coli (Kumar et al., 2014). Large-scale production of rhIFN-g has been recently reported by using prokaryotic E.coli expression system in fed-batch culture (Babaeipour et al 2007). In most cases, over-expression of rhIFN-g resulted in the formation of inclusion body in the cytoplasm (Kumar et al., 2014). The importance of IFN- in antiviral response against NDV in chicken fibroblasts has been demonstrated recently (Yang et al., 2020).
Among the most potent solubility enhancing proteins characterized to date are the E. coli maltose binding protein, MBP (40 kDa) and N-utilizing substance A, NusA (54.8 kDa). MBP and NusA act as solubility enhancing partners and are especially suited for the expression of proteins prone to form inclusion bodies (Sorensen and Mortensen, 2005). MBP fused with mouse leukemia inhibitory factor (mLIF) was used for its soluble expression in the cytoplasm of E.coli (Guo et al., 2020).
Soluble human fibroblast growth factor 21 (hFGF21), and human oncostatin M(OSM) were also expressed in E. coli by MBP-tagging (Nguyen et al., 2016, 2019a). Both MBP and NusA have been used for the solubilization of highly insoluble ScFv antibodies in the cytoplasm of E. coli (Bach et al., 2001). Recently, a comparative expression study showed that solubility was enhanced by using MBP and NusA fusion constructs at lower temperature of 18 °C (Nguyen et al., 2017).
The export of hGM-CSF to the periplasm using the pelB leader sequence marginally increased hGM-CSF expression; however fusion with a MBP-tag resulted in the maximum expression of ~70 mg/ml (Bhattacharya et al., 2005). It has also been reported that the native endoxylanase signal sequence is efficient in secreting recombinant proteins to the culture supernatant (Srivastava and Mukherjee, 2001) or to the periplasm (Jeong and Lee, 2001). The pelB signal sequence of pectate lyase B from Erwinia carotovora is commercially available and commonly used for periplasmic export of recombinant proteins. Several studies have employed the pelB signal sequence for secretion of recombinant product in bacterial host (Cho et al., 2018; Santos et al., 2019; Zhou et al., 2019; Perez-Perez et al., 2020).
Asparaginase and endoxylanse signals have been shown to improve hGM-CSF expression in E.coli (Khasa et al., 2011). The asparaginase signal sequence also helped in efficient secretion of soluble asparaginase to the extracellular space (Khushoo et al., 2005). Human Interferon-gamma has been chosen mainly due to its intrinsic nature to easily form inclusion bodies and also high level of expression. The main objective of this study is to compare the expression level of different rhIFN-g constructs as fusion proteins with nusA, endoxylanase signal, pelB leader and asparginase signal sequences. The expression level of target protein may reveal the regulatory role and influence of different fusion partners in host cells. This will be useful for selection of best host-vector combinations and other optimization studies in recombinant protein expression using E.coli.
MATERIALS AND METHODS
Bacterial strains and Plasmids: E.coli BL21(DE3) (Stratagene, USA) strain was used as host for expression of rhIFN-g and fusion rhIFN-g genes. E.coli DH5a (Amersham, USA) was used for cloning purposes and maintenance of plasmids. The plasmids employed were pRSET-A (Invitrogen), pET22b, pET14b (Novagen, USA) and pET 43a (+) (Novagen, USA).
Cloning of rhIFN-g gene: Primers bearing NdeI and BamHI restriction sites were used to amplify rhIFN-g cDNA by RT-PCR method from total RNA isolated from peripheral blood mononuclear cells as described previously (Vaiphei et al., 2009). The amplified cDNA fragment was cloned between the NdeI and BamHI sites of plasmid pET14b (Novagen) and pRSET-A (Invitrogen) to obtain the plasmid pET14-IFNg and pRSET-IFNg respectively.
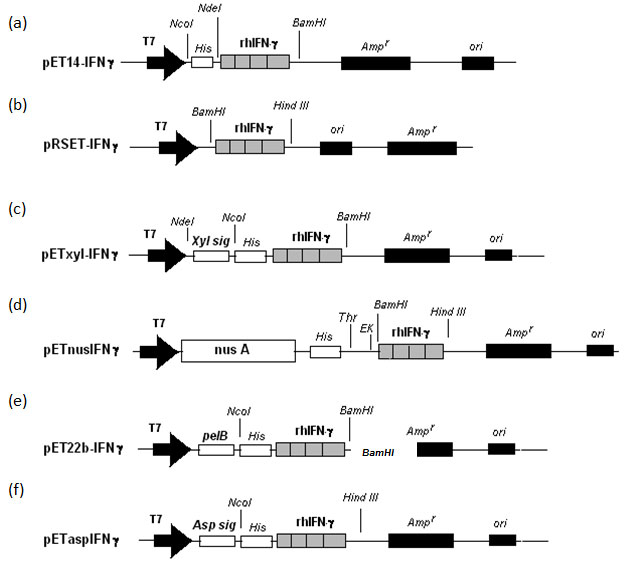
Construction of fusion plasmids: The plasmid pETnusAIFNg (hIFN-g gene fused with the nusA gene) was constructed by amplifying the hIFN-g fragment from the plasmid pRSET-IFNg and then introducing it into another plasmid pET-43a (Figure 1). For this, two primers were used in which the forward primer 5’CGC GGATCC CAG GAC CCA TAT GTA 3’ was designed to contain a BamHI restriction site and the reverse 5’ CCC AAGCTT TTA CTG GGA TGC TCT 3’ have Hind III restriction enzyme site. This allowed the fusion of rhIFN-g gene at its N-terminal with the nusA gene (Figure1d). The plasmid pETxyl-IFNg (rhIFN-g gene cloned under the endoxylanase signal sequence) was constructed by inserting the rhIFN-g gene fragment in plasmid PET-xyl (Srivastava and Mukherjee, 2001).
Two restriction enzymes NcoI and BamHI were used to double digest the pET14-IFNg to retrive the rhIFN-g gene fragment and ligated into pET-xyl vector digested with the same restriction enzymes (Figure1c). Expression vector for rhIFN-g fused with pelB leader sequence was constructed by inserting the NcoI and BamHI digested rhIFN-g fragment in pET22b vector also digested with the same restriction enzymes resulting in pET22-IFNg (Figure 1e). Also, for the construction of expression vector pETasp-IFNg (rhIFN-g gene cloned under the asparginase signal sequence), NcoI and BamHI fragment of rhIFN-g from pET14IFNg was ligated to NcoI/BamHI digested pET22asp vector as shown in Figure 1f (Khushoo et al., 2005).
Expression and Quantification of rhIFN-g as fusion protein: BL-21(DE3) cells containing rhIFN-g plasmid constructs from Figure 1 were grown in 5 ml LB media containing ampicillin (100mg/ml) and grown overnight at 37 oC shaker. About 1-2% of the overnight culture was transferred to 50 ml LB in 500 ml flask and grown at 37oC until OD600 reached 0.6. Then, the cells were induced with 1 mM IPTG and allowed to grow for 4 hours for expression of the recombinant proteins. Samples were collected at regular intervals for SDS-PAGE analysis and rhIFN-g production was quantified as described earlier (Vaiphei et al., 2009). In order to distinguish between soluble and insoluble protein fractions, the pellets were resuspended in phosphate buffer and incubated at 37 oC for 15 minutes in the presence of 200 mg/ml lysozyme. The samples were then lysed by sonication. This was followed by centrifugation at 14,000 rpm for 20 minutes, which pelleted the cell debris and any insoluble protein fraction. The supernatant consisting of the soluble cytoplasmic fraction was analysed. The insoluble pellets were resuspended in PBS containing 2% SDS in order to solubilize the inclusion bodies with appropriate dilution. The specific product yield was calculated for determining the rhIFN-g production as described earlier (Vaiphei et al., 2009).
RESULTS AND DISCUSSION
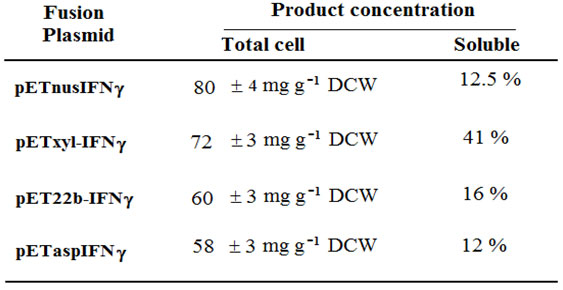
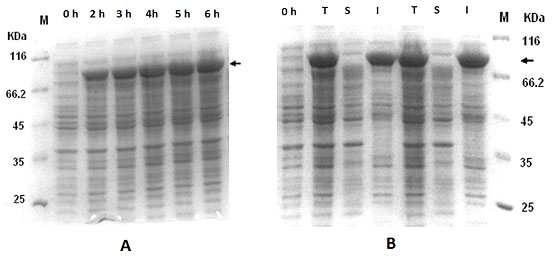
Expression of IFN-g-nusA fusion protein:Expression of rhIFN-g and nusA fusion was checked by inducing cells containing pETnusIFNg for 5 hours. The SDS-PAGE profile shows an induced band slightly above 70 kD which corresponds to rhIFN-g and nusA fusion protein which comprised about 30 % of the total cellular protein (Figure 2) and more than 85 % of the fusion protein was in the form of inclusion bodies (Table 1). On the other hand, the expression level did not increase significantly even after the 5 hour post induction (Figure 2). Besides, there was no significant improvement of soluble protein expression at 30 oC (data not shown).
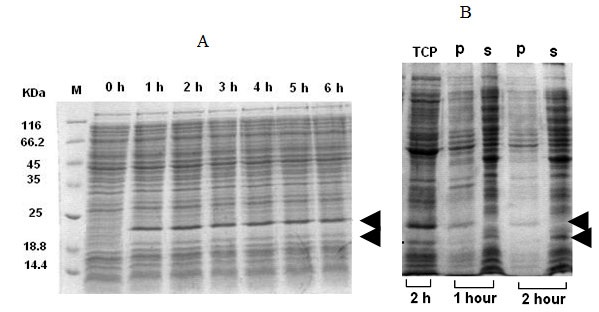
Expression of IFN-g-xylanase signal fusion:For the expression of rhIFN-g-xylanase signal fusion, pETxyl-IFNg containing cells were induced with IPTG for several hours. In this case, the recombinant protein mostly occurred in the form of immature precursor about the size of 21 kD and a very low level of the processed form of 19 kD (Figure 3A). The mature form constitutes the soluble fraction whereas the insoluble fraction comprised of the immature protein (Figure 3B). The maximum specific product yield of soluble rhIFN-g was found to be approximately 30 mg g-1 DCW that is about 41 % of the total cellular fraction (Table 1).
Expression of IFN-g-pelB leader fusion:The plasmid pET22b-IFNg was used for the expression of rhIFN-g and pelB leader sequence in BL-21 (DE3) cells. The expression level of the fusion protein was also found to be considerably low as seen from the SDS-PAGE and the pattern of expression suggested that the level of productivity remained the same till 4 hour after induction (data not shown). The major part of the insoluble fraction constitutes the recombinant protein being produced as fusion partner. The specific product yield of the soluble form was estimated to be about 16 % of the total fraction (Table 1).
Expression of IFN-g-asparginase signal fusion:The fusion rhIFN-g with the asparginase signal showed that majority of the recombinant fusion protein was synthesized in insoluble form (Table 1). The maximum specific product yield estimated being from total cell fraction was approximately 58 mg g-1 DCW from (Table 1). The expression level at 2 hour and 3 hour post induction time did not showed comparable difference (Data not shown).
Table 1. Comparison of different IFNg fusion proteins expression
Figure 1: Construction of different fusion constructs for this study.
Figure 2: Expression of IFNg and nusA fusion protein (pETnusIFNg) in E.coli. (A) Comparison of expression level at different post-induction time intervals. (B) Expression profiles showing total cell lysate (T), soluble fraction (S) and insoluble fraction (I).
Figure 3: Expression of IFNg and endoxylanase signal sequence fusion protein (pETxyl-IFNg) at different post-induction time intervals (A). Expression profile at 2h post-induction of total cell-lysate , insoluble fraction (p) and soluble fraction (s).
The solubility of the recombinant protein rely on several factors such as lowering the rate of protein synthesis by using moderate or weak promoters, substitution of the amino acid residues (Dale et al., 1994) or employing low temperature (Schein, 1993; Nguyen et al., 2017). However, the strategy involving the substitution of amino acid residues is limited to applications where these substitutions do not affect the desired function or stability of the target protein. Another well-known approach is fusion of the target proteins to highly soluble partners to increase the overall solubility of the fusion protein (LaVallie et al., 1993; Wilkinson et al., 1995), ubiquitin (Baker, 1996; Pilon et al., 1996); NusA (Davis et al., 1999). Unlike the affinity tags, solubility tags can differentially affect the target protein expression and therefore, heterologous expression systems involving fusion tags for improving solubility have continued to be tested (Costa et al., 2013; Nguyen et al., 2019b; Ki and Pack, 2020).
The propensity for rhIFN-g like proteins to form insoluble aggregates in E.coli has been well known. In this study, rhIFNg-nusA fusion showed highest total protein expression level as compared to the other fusion partners. There was no significant difference in the expression level at 30 oC and 37 oC. Large fusion tags like NusA exhibit chaperon-like activities that slow down translation process providing more time for protein folding and stabilization of target protein (Costa et al., 2014). The rhIFNg-xyl fusion showed both soluble and insoluble in the form of matured and immatured respectively (Figure 3B). This also indicates that the processed or matured form were soluble whereas the unprocessed ones are insoluble. However, this signal peptide was by far the most effective for expression of the fusion protein in soluble form.
Similarly, previous report also showed that native endoxylanase signal sequence fusion helped in efficient secretion of recombinant proteins to the culture supernatant (Srivastava and Mukherjee, 2001). In this work, pelB leader and asparaginase fusion partners have no significant increased in the level of soluble rhIFN-g (Table 1). The use of pelB leader sequence was shown earlier to have only marginal increase of hGM-CSF in the periplasmic space (Bhattacharya et al., 2005). In spite of using genetic strategy of fusion with different soluble partners, rhIFN-g was produced primarily as inclusion bodies demonstrating the propensity of the protein to aggregate upon over-expression in E.coli as host. This could possibly prevent efficient export to the periplasm and/or extracellular space which led to a drastic reduction in product yields. Since different fusion tags can have variable affect on the target protein expression, optimization with different combinations may be required on trial-and-error basis for best results (Costa et al., 2013, 2014; Paraskevopoulou and Falcone, 2018; Nguyen et al., 2019b; Ki and Pack, 2020).
Moreover, there is no direct correlation between the propensity of inclusion body formation of a certain protein and its intrinsic properties, such as molecular weight, hydrophobicity, folding pathways, and so on (Villaverde and Carrio, 2003). The reducing environment provided by the cytosol is suitable for the formation of inclusion body only in the case of disulfide bonded proteins, however, rhIFN-g do not contain cysteine residues for such bonding. Moreover, if the expressed protein is toxic to the host, the formation of inactive inclusion bodies might increase the viability of the cells and the yield of the target protein (Seo et al., 2005; Haught et al., 1998). It is obvious that the cellular pathway would prefer the inclusion bodies or non-production of the recombinant protein as seen from this study, to maintain cellular viability. Low or non expression of rhIFN-g with certain fusion partners could be the result of transcriptional or translational regulation of the bacterial genome. The cellular machinery might have adopted an alternative means to degrade or cleave the newly synthesized polypeptides to avoid formation of a soluble protein that could be lethal for the cells.
ACKNOWLEDGEMENTS
CSIR for funding and KJ lab members
Conflicts of interest
The authors declare no conflict of interest
REFERENCES
Babaeipour V, Shojaosadati SA, Robatjazi SM, et al. (2007) Over-production of human interferon-g by HCDC of recombinant Escherichia coli. Process Biochemistry 42:112–117.
Bach H, MazorY, Shaky S, et al. (2001) Escherichia coli maltose-binding protein as a molecular chaperone for recombinant intracellular cytoplasmic single-chain antibodies. J Mol Biol. 312(1): 79-93.
Baker RT. (1996) Protein expression using ubiquitin fusion and cleavage. Curr Opin Biotechnol. 7(5): p. 541-6.
Bhattacharya P, Pandey G, Srivastava P, et al. (2005) Combined effect of protein fusion and signal sequence greatly enhances the production of recombinant human GM-CSF in Escherichia coli. Mol Biotechnol. 30:103-16.
Costa SJ, Almeida A, Castro A, et al. (2013) The novel Fh8 and H fusion partners for soluble protein expression in Escherichia coli: a comparison with the traditional gene fusion technology. Appl Microbiol Biotechnol 97:6779–6791.
Costa S, Almeida A, Castro A, et al. (2014) Fusion tags for protein solubility, purification and immunogenicity in Escherichia coli: the novel Fh8 system. Front Microbiol 5:63.
Cho YH, Kim SJ, Kim JY, et al. (2018) Effect of PelB signal sequences on Pfe1 expression and ω-hydroxyundec-9-enoic acid biotransformation in recombinant Escherichia coli. Appl Microbiol Biotechnol. 102(17):7407-7416.
Dale GE, Broger C, Langen H, et al. (1994) Improving protein solubility through rationally designed amino acid replacements: solubilization of the trimethoprim-resistant type S1 dihydrofolate reductase. Protein Eng. 7(7): p. 933-9.
Davis GD, Elisee C, Newham DM et al. (1999) New fusion protein systems designed to give soluble expression in Escherichia coli. Biotechnol Bioeng. 65(4): p. 382-8.
Guo Y, Yu M, Jing N, et al. (2018) Production of soluble bioactive mouse leukemia inhibitory factor from Escherichia coli using MBP tag. Protein Expr Purif 150:86–91.
Haught C, Davis GD, Subramanian R, et al. (1998) Recombinant production and purification of novel antisense antimicrobial peptide in Escherichia coli. Biotechnol Bioeng. 57(1): 55-61.
Jeong KJ, Lee SY. (2001) Secretory production of human granulocyte colony-stimulating factor in Escherichia coli. Protein Expr Purif. 23:311-8.
Khasa YP, Khushoo A, Tapryal S, et al. (2011) Optimization of human granulocyte macrophage-colony stimulating factor (hGM-CSF) expression using asparaginase and xylanase gene’ssignal sequences in Escherichia coli. Appl Biochem Biotechnol. 165(2):523-37.
Khushoo A, Y Pal, Mukherjee KJ. (2005) Optimization of extracellular production of recombinant asparaginase in Escherichia coli in shake-flask and bioreactor. Appl Microbiol Biotechnol 68:189-97.
Ki MR, Pack SP. (2020) Fusion tags to enhance heterologous protein expression. Appl Microbiol Biotechnol. 104(6):2411-2425.
Kim TW, Chung BH, Chang YK. (2005) Production of soluble human interleukin-6 in cytoplasm by fed-batch culture of recombinant E. coli. Biotechnol Prog. 21(2): 524-31.
Kumar M, Singh M, Singh SB. (2014) Optimization of conditions for expression of recombinant interferon-γ in E.coli. Mol Biol Rep. 41: 6537–6543.
LaValli ER, DiBlasio EA, Kovacic S, et al. (1993) A thioredoxin gene fusion expression system that circumvents inclusion body formation in the E. coli cytoplasm. Biotechnology (N Y). 11(2):187-93.
Lilie H, Schwarz E, Rudolph R. (1998) Advances in refolding of proteins produced in E. coli. Curr Opin Biotechnol. 9(5):497-501.
Makrides SC. (1996) Strategies for achieving high-level expression of genes in Escherichia coli. Microbiol Rev. 60:512-38.
Mamane Y, Heylbroeck C, Genin P, et al. (1999) Interferon regulatory factors: the next generation. Gene. 237(1):1-14.
Nekoufar S, Fazeli A, Fazeli MR. (2020) Solubilization of Human Interferon β-1b Inclusion Body Proteins by Organic Solvents. Adv Pharm Bull. 10(2):233-238.
Nguyen AN, Song J-A, Nguyen MT, et al. (2017) Prokaryotic soluble expression and purification of bioactive human fibroblast growth factor 21 using maltose-binding protein. Sci Rep 7:16139.
Nguyen MT, Prima MJ, Song JA, et al. (2019a) Prokaryotic soluble overexpression and purification of oncostatin M using a fusion approach and genetically engineered E. coli strains. Sci Rep 9:13706.
Nguyen TKM, Ki MR, Son RG, et al. (2019b) The NT11, a novel fusion tag for enhancing protein expression in Escherichia coli. Appl Microbiol Biotechnol 103:2205–2216.
Paraskevopoulou V, Falcone F. (2018) Polyionic tags as enhancers of protein solubility in recombinant protein expression. Microorganisms 6: 47.
Perez-Perez DA, Pioquinto-Avila E, Arredondo-Espinoza E, et al. (2020) Engineered small metal-binding protein tag improves the production of recombinant human growth hormone in the periplasm of Escherichia coli. FEBS Open Bio. 10(4):546-551.
Santos BD, Morones-Ramirez JR, Balderas-Renteria I, et al. (2019) Optimizing Periplasmic Expression in Escherichia coli for the Production of Recombinant Proteins Tagged with the Small Metal-Binding Protein SmbP. Mol Biotechnol. 61(6):451-460.
Singh A, Upadhyay V, Panda AK. (2015) Solubilization and refolding of inclusion body proteins. Methods Mol Biol. 1258:283–291.
Singh A, Upadhyay V, Singh A, et al. (2020) Structure-Function Relationship of Inclusion Bodies of a Multimeric Protein. Front Microbiol. 11:876.
Pilon AL, Yost P, Chase TE, et al. (1996) High-level expression and efficient recovery of ubiquitin fusion proteins from Escherichia coli. Biotechnol Prog. 12(3): p. 331-7.
Schein CH. (1993) Solubility and secretability. Curr Opin Biotechnol. 4(4): p. 456-61.
Sen GC, Lengyel P. (1992) The interferon system. A bird’s eye view of its biochemistry. J Biol Chem. 267(8): 5017-20.
Seo JH, Yeo JS, Cha HJ. (2005) Baculoviral polyhedrin-Bacillus thuringiensis toxin fusion protein: A protein-based bio-insecticide expressed in Escherichia coli. Biotechnol Bioeng.
Sorensen HP, Mortensen KK. (2005) Soluble expression of recombinant proteins in the cytoplasm of Escherichia coli. Microb Cell Fact 4(1):1.
Srivastava P and Mukherjee KJ. (2001) Cloning, characterization, and expression of xylanase gene from Bacillus lyticus in Escherichia coli and Bacillus subtilis. Prep Biochem Biotechnol 31:389-400.
Vaiphei ST, Pandey G, Mukherjee KJ. (2009) Kinetic studies of recombinant human Interfereon-gamma expression in continuous cultures of E.coli. J Ind Microbiol Biotechnol. 36(12): 1453-1458.
Villaverde A, Carrio MM. (2003) Protein aggregation in recombinant bacteria: biological role of inclusion bodies. Biotechnol Lett 25(17): 1385-95.
Wilkinson DL, Ma NT, Haught C, et al. (1995) Purification by immobilized metal affinity chromatography of human atrial natriuretic peptide expressed in a novel thioredoxin fusion protein. Biotechnol Prog. 11(3): p. 265-9.
Yang X, Arslan M, Liu X, et al. (2020) IFN-γ establishes interferon-stimulated gene-mediated antiviral state against Newcastle disease virus in chicken fibroblasts. Acta Biochim Biophys Sin (Shanghai). 52(3):268-280.
Yang Z, Zhang L, Zhang Y, et al. (2011) Highly efficient production of soluble proteins from insoluble inclusion bodies by a two-step-denaturing and refolding method. PloS One 6(7): e22981.
Zhou Z, Li Q, Xu R, Wang B, Du G, Kang Z. (2019) Secretory expression of the rat aryl sulfotransferases IV with improved catalytic efficiency by molecular engineering. 3 Biotech. 9(6):246.