Biotechnology Department, College of Agriculture and Food Science, King Faisal University, PO Box 400 Al Hofuf, Al-Ahsa 31982 Saudi Arabia
Corresponding author email: falmuhanna@kfu.edu.sa
Article Publishing History
Received: 06/10/2019
Accepted After Revision: 30/11/2019
Endomycorrhizae have the ability to exploit organic sources while providing their host plant with nitrogen and phosphorus, but the mutualistic molecular mechanism by which this occurs is unclear. In a previous study, five uncharacterized root proteins were found to be significantly upregulated in endomycorrhiza-colonized sorghum, which showed an increase in biomass and element uptake. However, the functions of these proteins are unknown. Therefore, in the current study, the functions of these five proteins were explored using computational analysis. Since these root proteins were not identified in plant protein sequences, they were submitted to Blast to search for homologous fungal protein data. The possible functions of the resulting 406 homologous proteins were investigated by performing several analyses, namely, phylogenetic analysis, sequence similarity network (SSN) analysis, genome neighborhood network (GNN) analysis, and functional network identification. The results of analyzing the uncharacterized sorghum root proteins revealed three integral membrane proteins among them: an APC amino-acid permease, a transmembrane transporter activity protein, and an acid protease. These results emphasize the role of endomycorrhiza–plant interactions in increased root permeability, resulting in the enhanced exchange of reciprocal resources in a symbiotic process.
Arbuscular Mycorrhiza, Mutualism, Endomycorrhiza, Sorghum, Roots, Membrane Proteins
Dhawi F. Endomycorrhizae Enhances Reciprocal Resource Exchange Via Membrane Protein Induction. Biosc.Biotech.Res.Comm. 2019;12(4).
Dhawi F. Endomycorrhizae Enhances Reciprocal Resource Exchange Via Membrane Protein Induction. Biosc.Biotech.Res.Comm. 2019;12(4). Available from: https://bit.ly/36cCKoQ
Copyright © Dhawi This is an open access article distributed under the terms of the Creative Commons Attribution License (CC-BY) https://creativecommons.org/licenses/by/4.0/, which permits unrestricted use distribution and reproduction in any medium, provide the original author and source are credited.
INTRODUCTION
The health of the biosphere, which largely depends on plant and environmental factors, shapes the interactions among dynamic communities involving macro- and microorganisms, such as bacteria and mycorrhizas. An arbuscular mycorrhiza (AM), also known as endomycorrhiza, colonizes and branches within the root cells of plants and stores oil-rich products in its vesicles. After successful colonization, the AM grows a branch-like structure (known as hyphae) that extends outside the roots for several centimeters in the soil. Hyphae facilitate nutrient uptake, particularly phosphate ions and nitrogen, in a tradeoff for carbon extraction from plant roots (Coleman et al., 2017). The beneficial effects that the AM confers to the plant occur across diverged membrane interfaces in a symbiotic relationship between the fungus and the host plant (Casieri et al., 2013). The development of AM colonization requires the plant cell’s secretory mechanism to improve the rapid branching of fungal hyphae and generate a periarbuscular membrane (Ivanov et al., 2019).
Extensive studies have focused on the influence of AMs on plant growth and biomass, nutrient availability, and metabolites (Dhawi et al., 2015; Dhawi et al., 2016; Dhawi and Hess, 2017a, b; Dhawi et al., 2018).The use of an AM alone or in combination with plant-growth-promoting bacteria was found to enhance the activity of the radical scavenging system in sorghum roots (Dhawi et al., 2017) and contribute to the upregulation of the metabolites associated with amino acid synthesis in foxtail millet (Dhawi et al., 2018). Many studies have suggested that the symbiotic process involves rhizobiome assembly governed by novel protein families (Sasse et al., 2018). One study reported the upregulation of 82 proteins in the plasma membrane of plants in response to mycorrhizal colonization (Aloui et al., 2018); thus, the plant’s carbohydrate and nitrogen‐derived transporters were enhanced during AM colonization (Hacquard et al., 2013). Although previous studies have provided insights into the system of rhizosphere microbiomes, little is known about plants’ molecular changes during colonization. Here, we reveal the function of five uncharacterized sorghum proteins in roots treated with endomycorrhiza. In a previous study, endomycorrhiza colonization resulted in an increase in sorghum root biomass and element uptake, together with a significant upregulation of five proteins with unknown functions (Dhawi et al., 2017).
In this study, the functions of these unknown proteins were explored using computational analysis. Several steps were taken to identify the functions of these proteins. The sequences of the uncharacterized sorghum root proteins were submitted to Blast to search for homologous protein sequences in fungal protein data since they were not identified in plant protein sequences. This resulted in the identification of 406 homologous proteins, which were then subjected to several analyses to determine their possible roles. These analyses included sequence similarity network (SSN) and genome neighborhood network (GNN) construction, as well as functional network identification. The results of analyzing the uncharacterized proteins revealed three integral membrane proteins: APC amino-acid permease, transmembrane transporter activity protein, and acid protease. These findings add to our understanding of the molecular mechanisms by which mycorrhizal synergetic activity enhances plant productivity.
MATERIAL AND METHODS
Plant growth conditions and treatments:Two-day-old germinated seedlings of Sorghum bicolor (BTx623, USDA) were planted in 600 g of pasteurized soil mix consisting of coarse sand and loam soil (1:1). The soil mix was inoculated with 9 g (~1320 propagules) of an endomycorrhizal mix (MycoApply® Endo, Valentine Country Inc, USA), and the control group was inoculated with 9 g of a pasteurized endomycorrhizal mix at 70 °C (Dhawi et al., 2015). The endomycorrhizal mix consisted of ~1320 spores of Glomus intraradices, G. mosseae, G. aggregatum, and G. etunicatum. Sorghum roots were harvested 90 days after treatment and subjected to protein extraction and identification. Protein was extracted from frozen roots. Three samples from each group were processed following the procedure described in Dhawi et al. (2017) and modified in Fukao et al. (2011).
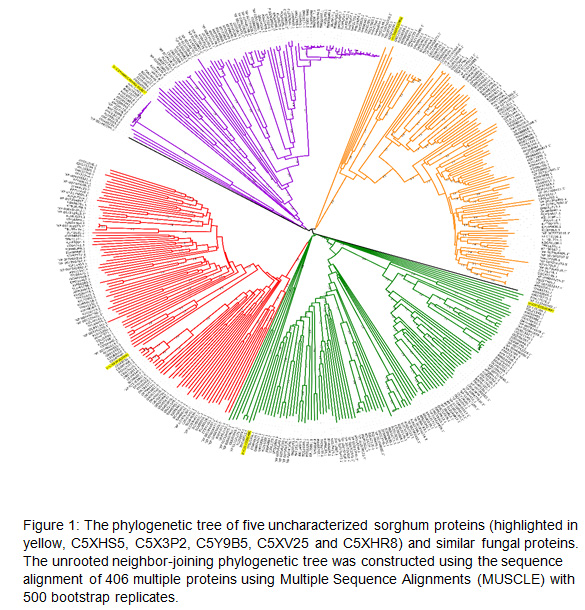
Protein processing of uncharacterized sorghum roots: The sequences of the five uncharacterized sorghum root proteins with increased fold changes (11–33-fold) (Table 1) were used to search for homologous fungal proteins using PSI Blast (https://blast.ncbi.nlm.nih.gov/Blast.cgi?PAGE=Proteins) (Altschul et al., 1997), which resulted in 401 protein sequences. These sequences were filtered using CD-HIT Suite: Biological Sequence Clustering and Comparison (http://weizhongli-lab.org/cdhit_suite/cgi-bin/index.cgi), with a 90% identity cut-off. The 90% cut-off step resulted in 406 protein sequences for performing a multiple sequence alignment (MSA). The MSA was created using the MEGA7 (Molecular Evolutionary Genetics Analysis) software version 6 (Tamura et al., 2013). The phylogenetic tree was established in MEGA6 and aligned using the Multiple Sequence Alignments (MUSCLE) program (Edgar, 2004) for 406 proteins, five of which were the uncharacterized upregulated sorghum root proteins. Protein sequences were aligned using the neighbor-joining method (Saitou and Nei, 1987) and UPGMA (unweighted pair group method), with 500 bootstrap replications for the phylogenetic assessment. The interactive tree of life (iTol) online web tool (http://itol.embl.deg) was then used to display and modify the tree colors (Letunic and Bork, 2016).
Table 1: Fold changes in uncharacterized sorghum proteins influenced by endomycorrhiza.
| Protein ID | Fold change compared with control |
| C5XHS5 | 33 |
| C5X3P2 | 20 |
| C5Y9B5 | 16 |
| C5XV25 | 13 |
| C5XHR8 | 11 |
Protein functional network and family assignment: A combination of different approaches was used to assign homologous proteins to protein families (Pfam) with putative functions. The 406 protein sequences consisted of uncharacterized sorghum root proteins and similar fungal protein sequences used to find their functions on the basis of their genomic, protein, and gene interactions using the sequence similarity network (SSN) and genome neighborhood network (GNN) approaches. The GNN approach was performed using the Enzyme Function Initiative-Enzyme Similarity Tool (EFI-EST) (https://efi.igb.illinois.edu/efi-est/; Gerlt et al., 2015). For EFI-EST, the 406 proteins were first converted to UniProt IDs. Only 60 proteins were identified in the UniProt database, and 346 were unidentified fungal proteins. The EFI-EST website was used to generate an SSN, and the resulting files were visualized using the Cytoscape software (Shannon et al., 2003). Sorghum protein sequences were screened for motifs using the MEME (Multiple EM for Motif Elicitation) tool (http://meme-suite.org/tools/meme) (Tanaka et al., 2011), with the detection level set to a maximum of eight motifs and an E-value equal to or less than 0.05.
RESULTS AND DISCUSSION
Phylogenetic tree analysis: The phylogenetic tree of 406 protein sequences contained five uncharacterized sorghum proteins and 401 similar fungal proteins aligned using Multiple Sequence Alignments (MUSCLE) with 500 bootstrap. The resulting unrooted phylogenetic tree consisted of 406 nodes, in which sorghum uncharacterized protein sequences clustered in five of them (Fig.1). Phylogenetic tree analysis identified uncharacterized sorghum protein C5XHS5 clustered in one clade with fungal protein of ORZ25315.1, which belonged to Amino acid/polyamine transporter I. However, sorghum protein C5Y9B5 was clustered with four fungal proteins, namely ‘XP 013245342.1, EDK37078.2, TDL21399.1 and OAX33366.1. C5XHR8 clustered with AWI66956.1 and CEI93594.1 On the other hand, two sorghum proteins were sorted with no cluster, with fungal proteins C5XV25 and C5X3P2 (Fig.1). The sorghum uncharacterized root protein C5XHS5 which was upregulated to 33 fold and showed to be close to fungal protein ORZ25315.1 based on phylogenetic analysis.
This protein belongs to the Absidia repens protein that functions as
transmembrane transporter activity (https://www.uniprot.org/uniprot/A0A1X2J0L6). The second sorghum protein was C5Y9B5, which showed to be close to all these fungal proteins :
XP 013245342.1, which belongs to Tilletiaria anomala that functions as Acid protease (https://www.uniprot.org/uniprot/A0A066WIX7); EDK37078.2, which belongs to uncharacterized protein of yeast Meyerozyma guilliermondii (https://www.uniprot.org/taxonomy/294746); TDL21399.1, which belongs to uncharacterized protein of fungus Caballeronia sordidicola (https://www.uniprot.org/uniprot/A0A4R1P5H9) and close to OAX33366.1 protein of fungus of Rhizopogon vinicolor, which functions as Acid protease (https://www.uniprot.org/taxonomy/1314800). Furthermore, C5XHR8 clustered with fungal protein of AWI66956.1 belongs to fungal proteins of Neocallimastix cameroonii and known as the Glycosyl hydrolase family 17 (https://www.uniprot.org/uniprot/A0A2S1TZ75) and with CEI93594.1 belongs to uncharacterized protein of fungus Caballeronia sordidicola (https://www.uniprot.org/uniprot/A0A0A1NM33).
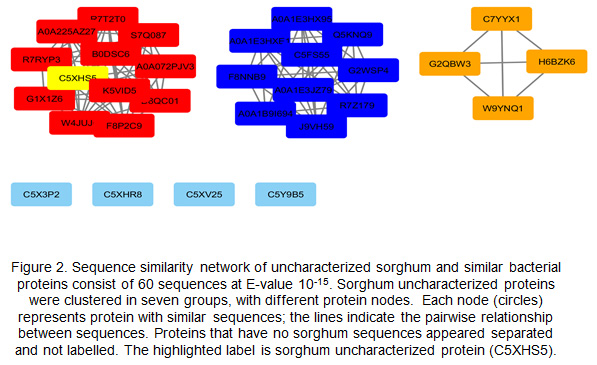
Sequence similarity network (SSN):The SSN is used to analyze 60 sequences containing five sorghum uncharacterized protein sequences and 55 fungal protein sequences. These sequences were obtained by PSI-BLAST at E.value 10-15; the sequences were clustered based on their similarities in four clusters. The SSN clusters contained one uncharacterized sorghum protein, whereas, the other four proteins were separated with no clusters due to a different E.value score. These resulting SSNs were used to establish Genome Neighborhood Network (Fig. 2).
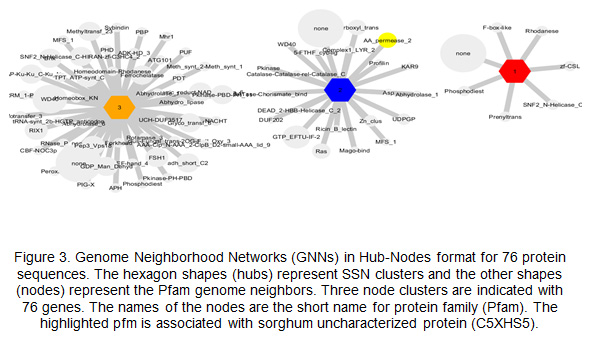
Genome Neighborhood Networks GNN, and functional partners: The SNN results are used to build GNNs. The output of GNN in Hub-Nodes format showed the number of Pfam gene neighbors that have been found in each cluster of sequences. The SNNs of uncharacterized protein sequences of sorghum were clustered in seven nodes. The nodes were protein families, arranged based on their genomic context and shared 86 protein families (Fig.3). The resulting SSNs were used to establish Genome Neighborhood Network identified nodes of protein families, arranged based on their genomic context. The protein family identified showed several forms in KEGG, namely: (https://www.genome.jp/dbget-bin/www_bget?lbc:LACBIDRAFT_191715) ,
(https://www.genome.jp/dbget-bin/www_bget?gtr:GLOTRDRAFT_140033) and
(https://www.genome.jp/dbget-bin/www_bget?hir:HETIRDRAFT_66031). On the other hand, the analysis of uncharacterized sorghum root proteins identified one protein family, which was Amino acid permease (Pfm: PF13520) (Fig. 3).
The nature of the mutualistic interaction between plants and endomycorrhiza has been extensively studied. However, endomycorrhiza symbiotic proteins and signaling molecules have remained unexplored. In the current study, the uncharacterized sorghum root protein C5XHS5, which was upregulated by 33-fold, was shown to be close to fungal protein ORZ25315.1—an Absidia repens protein that has transmembrane transporter activity. C5Y9B5 was shown to be close to two fungal proteins: one belongs to Tilletiaria anomala, and the other protein belongs to the fungus Rhizopogon vinicolor. Both of these proteins function as an acid protease. C5XHR8 clustered with the fungal protein of Neocallimastix cameroonii known as glycosyl hydrolase family 17. One uncharacterized sorghum protein was grouped in SSN clusters, whereas the other four proteins were separated without clusters because of their different E-value scores. The only uncharacterized sorghum protein that was identified was C5XHS5. This protein was especially upregulated (33-fold) in sorghum roots after endomycorrhizal colonization, and it clustered with 11 fungal proteins. Next, the resulting SSNs were used to establish a genome neighborhood network, which identified one protein family, namely, APC amino-acid permease.
Arbuscular mycorrhizal (AM) symbiosis, which is assumed to enhance water and nutrient absorption, is associated with massive arrays of membrane tubules that form between the plant protoplast and the cell wall (Ivanov et al., 2019). The plasma membrane-associated proteins that respond to mycorrhization support the host plant’s control of sugar uptake and mediate the replacement of phospholipids by phosphorus-free lipids in the plasmalemma of mycorrhizal roots (Aloui et al., 2018). This explains the increase in carbohydrate and nitrogen‐derived transporters during the endomycorrhiza’s symbiotic stages since these transporters enhance the transfer of reciprocal resources between the host and the symbiont (Hacquard et al., 2013).
The uncharacterized sorghum root proteins that were highly upregulated are part of the protein families that function as a transmembrane transporter, acid protease, and APC amino-acid permease. This result is similar to that of a previous study on amino-acid permease that described this protein as a molecular tool of endomycorrhiza (Glomus mosseae). This study identified an increase in amino-acid permease following exposure to organic nitrogen from the soil, thereby enhancing amino acid acquisition (Cappellazzo et al., 2008). The amino acid transporter (AAT) gene (LjLHT1.2) that encodes for the lysine–histidine–transporter (LHT)-type amino acid transporter induced in mycorrhizas is involved in the complex mechanisms of amino acid recycling in colonized roots (Guether et al., 2011).
Conclusion: The analyses of five significantly upregulated proteins in endomycorrhiza-colonized sorghum roots add to our understanding of the complex molecular mechanisms of mycorrhizal synergetic activity. The enhancement of plant productivity is the result of the mycorrhizal-induced upregulation of plant membrane proteins and the expression of the associated genes, thereby increasing root permeability and reciprocal resource exchange between the host and the symbiont.
REFERENCES
Aloui, A., Recorbet, G., Lemaître-Guillier, C., Mounier, A., Balliau, T., Zivy, M. & Dumas-Gaudot, E. (2018). The plasma membrane proteome of Medicago truncatula roots as modified by arbuscular mycorrhizal symbiosis. Mycorrhiza, 28(1), 1-16.
Altschul, S. F., Madden, T. L., Schäffer, A. A., Zhang, J., Zhang, Z., Miller, W., & Lipman, D. J. (1997). Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic acids research, 25(17), 3389-3402.
Cappellazzo, G., Lanfranco, L., Fitz, M., Wipf, D., & Bonfante, P. (2008). Characterization of an amino acid permease from the endomycorrhizal fungus Glomus mosseae. Plant Physiology, 147(1), 429-437.
Casieri, L., Lahmidi, N. A., Doidy, J., Veneault-Fourrey, C., Migeon, A., Bonneau, L., … & Brun, A. (2013). Biotrophic transportome in mutualistic plant–fungal interactions. Mycorrhiza, 23(8), 597-625.
Coleman, D. C., Callaham, M. A., & Crossley Jr, D. A. (2017). Fundamentals of soil ecology. Academic press.
Dhawi, F., Datta, R., & Ramakrishna, W. (2018). Metabolomics, biomass and lignocellulosic total sugars analysis in foxtail millet (Setaria italica) inoculated with different combinations of plant growth promoting bacteria and mycorrhiza. Communication in Plant Sciences, 8, 8-14.
Dhawi, F., & Hess, A. (2017a). Poor-Soil Rhizosphere Enriched with Different Microbial Activities Influence the Availability of Base Elements. Open Journal of Ecology, 7(08), 495.
Dhawi, F., & Hess, A. (2017b). Plant Growth-Prompting Bacteria Influenced Metabolites of Zea mays var. amylacea and Pennisetum americanum p. in a Species-Specific Manner. Advances in Biological Chemistry, 7(05), 161.
Dhawi, F., Datta, R., & Ramakrishna, W. (2017). Proteomics provides insights into biological pathways altered by plant growth promoting bacteria and arbuscular mycorrhiza in sorghum grown in marginal soil. Biochimica et Biophysica Acta (BBA)-Proteins and Proteomics, 1865(2), 243-251.
Dhawi, F., Datta, R., & Ramakrishna, W. (2015). Mycorrhiza and PGPB modulate maize biomass, nutrient uptake and metabolic pathways in maize grown in mining-impacted soil. Plant Physiology and Biochemistry, 97, 390-399.
Fukao, Y., Ferjani, A., Tomioka, R., Nagasaki, N., Kurata, R., Nishimori, Y. & Maeshima, M. (2011). iTRAQ analysis reveals mechanisms of growth defects due to excess zinc in Arabidopsis. Plant Physiology, 155(4), 1893-1907.
Guether, M., Volpe, V., Balestrini, R., Requena, N., Wipf, D., & Bonfante, P. (2011). LjLHT1. 2—a mycorrhiza-inducible plant amino acid transporter from Lotus japonicus. Biology and fertility of soils, 47(8), 925.
Hacquard, S., Tisserant, E., Brun, A., Legué, V., Martin, F., & Kohler, A. (2013). Laser microdissection and microarray analysis of Tuber melanosporum ectomycorrhizas reveal functional heterogeneity between mantle and H artig net compartments. Environmental Microbiology, 15(6), 1853-1869.
Ivanov, S., Austin, J., Berg, R. H., & Harrison, M. J. (2019). Extensive membrane systems at the host–arbuscular mycorrhizal fungus interface. Nature plants, 5(2), 194.
Letunic, I., & Bork, P. (2016). Interactive tree of life (iTOL) v3: an online tool for the display and annotation of phylogenetic and other trees. Nucleic acids research, 44(W1), W242-W245.
Saitou, N., & Nei, M. (1987). The neighbor-joining method: a new method for reconstructing phylogenetic trees. Molecular biology and evolution, 4(4), 406-425.
Sasse, J., Martinoia, E., & Northen, T. (2018). Feed your friends: do plant exudates shape the root microbiome?. Trends in plant science, 23(1), 25-41.
Tamura, K., Stecher, G., Peterson, D., Filipski, A., & Kumar, S. (2013). MEGA6: molecular evolutionary genetics analysis version 6.0. Molecular biology and evolution, 30(12), 2725-2729.