1Department of Biotechnology, Barkatullah University, Bhopal-462026
2Department of Biotechnology, Faculty of Science, RKDF University, Bhopal-462036
Corresponding author Email: ankita.verma1234@gmail.com
Article Publishing History
Received: 19/09/2018
Accepted After Revision: 23/11/2018
Antagonistic endophytic fungal community resident in medicinal plant Stevia rebaudiana Bertoni was studied at two sites within Bhopal, M.P. Among 339 recovered endophytic isolates from foliar tissues, 40 fungal isolates were found antagonistic against Sclerotinia sclerotiorum, casual agent of disease stem rot in stevia and soybean (Glycine max.). Antagonistic fungal population (40 isolates) consisted of 52.5% Hyphomycetes and 2.5% each of Coleomycetes, Basidiomycetes, Ascomycetes and Sterile mycelia. The percent colonization frequency of antagonistic endophytic community in foliar tissues ranged from 0.3%-5.3% whereas percent dominance was of the order, ranged from 2.31%-40.8%. Diversity analysis of the antagonistic endophytic population was determined in terms of Shanon index, Simpson index, Species eveness, Menhinick and Margalef richmess index .Antagonistic endophytic population was also evaluated for IAA production, siderophore and phosphorus solubilisation, considered as plant growth promotory attributes. Identification of the antagonistic endophytes was carried out by rDNAsequencing of the ITS region.
Diversity Index, S. Sclerotiorum, Its Region, Antagonistic Endophytes, Sequence Phylogeny
Verma A, Saini S, Johri B. N, Prakash A. Diversity Analysis and Characterization of Antagonistic Endophytic Population from Stevia Rebaudiana. Biosc.Biotech.Res.Comm. 2018;11(4).
Verma A, Saini S, Johri B. N, Prakash A. Diversity Analysis and Characterization of Antagonistic Endophytic Population from Stevia Rebaudiana. Biosc.Biotech.Res.Comm. 2018;11(4). Available from: https://bit.ly/2mkEkDy
Introduction
Fungal endophytes possess huge diversity morphologically and biochemically (Strobel and Daisy, 2003). Endophytic fungi are known to reside in the tissues of plants above ground as well as below ground, parts of the plant (Zhang et al. 2006; Kusari et al. 2012). Endophytic fungi are an assemblage of microorganisms that chiefly belong to class Ascomycetes of kingdom fungi. A significant literature is available so far to show that these microorganisms, under laboratory culture conditions, produce numerous structurally diverse biologically active secondary metabolites that include antimicrobial substances. Different ecological factors such as seasonality, nearby vegetation and humidity influence the distribution of endophytic fungi in the host (Taylor et al; 1999; Toofanee and Dulymamode; 2002 Lumyong et al 2009, Dissanayake et al., 2016; Ratnaweera et al., 2017, Ratnaweera et al., 2018).
Stevia rebaudiana Bertoni is an herbaceous polyannual plant of the sunflower family (Fam. Asteraceae), generally known as candy leaf, sweet leaf, sweet leaf, or sugar leaf. The Stevia leaves also contain variety of glycosides compounds viz., flavonoid glycosides, Stevioside, Rebaudioside A, Rebaudioside C, coumarins, cinnamic acids, phenylpropanoids and some essential oils, (Midmore and Rank, 2002 Lavini et al., 2008).
Stevia rebaudiana Bert. Is a good source of sweeteners and is about 300 times sweeter than sucrose owing to presence of steviosides in its leaves. Previous studies have reported clearly the diversity of these fungal fungal endpphytes from Stevia rebaudiana which also indicates the presence of Alternaria,, Aspergillus, Monodictys, and Curvularia fungal genus from leaf of Stevia rebaudiana (Bert.). (Prakash et al., 2008; Madhumita and Chandra, 2013) Furthermore, these fungal isolates have been reported from almost all climatic regions of the globe viz., tropical, temperate and alpine (Arnold, 2007; Halmschlager et al 1993; Higgins et al 2007). The application of biocontrol agents has become one of the most promising tools for reducing the use of chemical pesticides in agriculture. The antagonism of biocontrol agent is based on different mechanisms i.e. nutrients, mycoparasitism, plant growth promotion and induction of the defense responses in plants (Howell, 2003, Sen
et al, 2012; Hamzah et al, 2018).
In the present investigation the endophytes recovered from Stevia rebaudiana leaves have been tested for antagonistic abilities against Sclerotinia sclerotiorum, which is the major phytopathogen affecting varieties of crop plants in Central India (Prakash et al., 2008, Verma et al 2004).
Therefore, present investigation was carried out to understand the generic diversity of endophytic fungi in leaves of Stevia rebaudiana Bertoni and to compare the antagonistic endophytic assemblages in samples collected from two different sites in the same region. Thus a specific rationale for the selection of stevia plant for endophyte isolation and natural product discovery is used.
Materials And Methods
Stevia rebaudiana Bertoni: Stevia rebaudiana Bertoni was selected as the target plant for isolation of fungal endophytes. Sampling was carried out from two sites. The first site was Misrod (23º16’N; 76º36’E), a village situated nearly 22 km away from the capital city Bhopal in the state of Madhya Pradesh. The second site of sampling was green house grown plants in the campus of (23º20’N; 77º45’E) Barkatullah University, Bhopal.
Samples Collection and Surface sterilization: Healthy and mature plants were carefully chosen for sampling and leaves were collected randomly. Plant material was brought to the laboratory in sterile bags. Surface sterilization of foliar tissues were done using reagents like 70% ethanol, 4% sodium hypochlorite and sterile distilled water for different time period for effective surface sterilization process. Leaves were thoroughly washed several times in sterile distilled water (SDW) for 5-10 sec then sterilized by exposing them to 70% ethanol for 2 min followed by treatment with 4% sodium hypochlorite for 2 min. The leaves were now immersed in sterile water (SDW) for 2-5 sec and allowed to dry on blotting paper.
Isolation of endophytic fungi from foliar tissues of plants: The surface sterilized leaf segments of 5mm size were placed on Potato dextrose Agar (Howksworth et al 1995), supplemented with chloramphenicol (0.2gl-1) to avoid bacterial contamination. Plates were incubated at 28±2 ºC in for 3-5 days and were observed regularly for fungal growth.
Analysis of data: The colonization frequency (% CF) of endophytic fungi was calculated according to Hata and Futai (1995) and dominance as per Kumaresan and Suryanarayanan (2002). Utilizing the data of percentage colonization of fungal endophytes in leaves, for two sites. Simpson’s Diversity indices and Shanon-Wiener indices were calculated. Species evenness and species richness was calculated according to Simpson (1949), Shanon and Weaver (1949), Ludwig & Reynolds (1988) and Margelef and Menhinick (1964)
In vitro antagonistic activity of fungal endophyte
Fungal isolates were screened for antagonism against Sclerotinia sclerotiorum by a dual culture technique on Potato Dextrose Medium (Szekeres et al, 2005).
Characterization of endophytes: The endophytic fungi were identified by their macroscopic & microscopic characteristics such as the morphology of the fruiting bodies and spore morphology. Morphological characterization was done on the basis of color, margin, reverse pigmentation & texture. (Rifai 1969).Antagonistic endophytic fungi are characterized functionally employing plate assays for amylase cellulase, Protease, pectinase, lipase and xylanase (Paterson & Bridge. 1994; Teather & Wood. 1982; Shakeri et al. 2007; Pointing. 1999; Sierra. 1957, Mishra et al, 2013, Aneja, 2003).
Plant growth promoting attributes: Plant growth promoting attributes of antagonistic endophyte were also studied. This included IAA (Brick et al. 1991) and siderophore production (Schwyn & Neilands. 1987) and phosphate solubilisation efficiency (Pikovskaya. 1948). Both qualitative and quantitative estimation were made.
Molecular identification of antagonistic endophyte: – Morphological identification of the organism was carried out at National Fungal Culture Collection of India (NFCCI), Agharkhar Research Institute, Pune. For molecular identification, total genomic DNA of the endophytic fungus was isolated directely from actively growing mycelium growing in Potato dextrose broth (PDB), using CTAB method (Sambrook and Russel, 2001). DNA amplification was performed by PCR using primer pair ITS1: TCCGTAGGTGAACCTGCGG and ITS4: TCCTCCGCTTGATATGC (White et al. 1990).PCR was carried out according to the following protocol: initial denaturation at 95 ºC for 5 min; denaturation at 95 ºC for 1 min; annealing at 55ºC for 45 sec; extension at 72 ºC for 10 min; steps 2-4 were repeated 35 times. Sequencing of PCR product was carried at Xcelris Labs Ltd, Ahmedabad. The sequenced data was subjected to BLAST algorithm and submitted to Genebank for accession number. The potential antagonistic endophytes were submitted at National Agriculturally Important Culture Collection (NAIMCC), culture collection facility at ICAR-NBAIM, Maunath Bhanjhan (U.P).
Phylogenetic analysis:To know the phylogenetic relationship among the isolates and also to confirm their taxonomical status, certain ITS rDNA sequences were chosen from GenBank databases via BLAST search analysis. The sequences were chosen from the top 20 database hits obtained in the blast search by querying the obtained sequences individually. These sequences were aligned using CLUSTAL W 1.83 (Thompson et al., 1994). Phylogenetic trees were generated by neighbourhood joining method with 100 bootstrapping replicates using MEGA version 5.
Results
Antagonistic Action and Diversity Analysis: A total of 339 recovered endophytic fungal isolates were screened for antagonistic ability against Sclerotinia sclerotiorum (culture obtained from Directorate of Soybean Research, Indore, M.P.) by using dual culture technique. The inhibition zone in dual plate assay averaged between 5 to 17 mm. Misrod field site harboured greater number of antagonists compared to the endophytes recovered from green house raised plants. Percent growth reduction of the pathogenic culture was recorded after 24 hrs. Isolate Aspergillus flavipes (NAIMCC-F-03153) strain 63 showed highest value of growth reduction i.e. 19% after 24 hrs followed by Alternaria alternata strain 99 and Aspergillus niger strain 89 (NAIMCC-F-03157) showed 17% and 18% growth reduction respectively (Fig 1); Least reduction of pathogen was recorded for isolate Alternaria brassicae strain 17 i.e. 6.9%. The percent growth reduction values ranged from 6.9% to 19%.
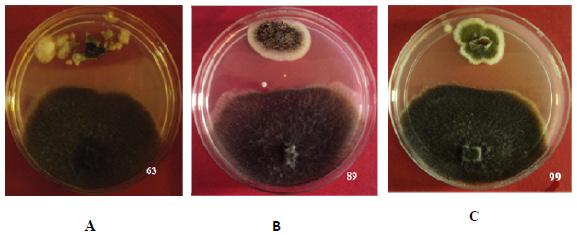 |
Figure 1: Dual culture assay shown by isolate A) Strain 63 B) Strain 89 C) Strain 99 |
A total of 40 endophytic fungal isolates which showed antagonistic activity against Sclerotinia sclerotiorum consisted of 52.5% Hyphomycetes followed by 2.5% Coleomycetes, Basidiomycetes, Ascomycetes and Sterile mycelia each. The percent colonization in tissues samples ranged from 1% – 36.6% (site 1) whereas 1%-10.6% (site 2) (Fig 2) and percentage dominance of antagonistic endophytes ranged from 1.1%-40.6% (site 1) and 4.3%-45.3% at site2 (Fig 3). Diversity analysis of the antagonistic population was carried out which showed significant diversity index values at site 1 as compared to site 2, whereas Margalef & Menhinick’s richness index value was maximum at site 2 as compared to site 1 (Table 1)
| Table 1: Diversity indices of two sites (Site 1 & Site 2) | ||
| Diversity indices | Site I | Site II |
| Simpson index (1-D) | 0.821 | 0.7982 |
| Shanon index (H’) | 1.856 | 1.781 |
| Eveness (EH/s) | 0.8001 | 0.8478 |
| Menhinick S/√N | 0.6305 | 1.121 |
| Margalef S-1/ln(n) | 1.378 | 1.638 |
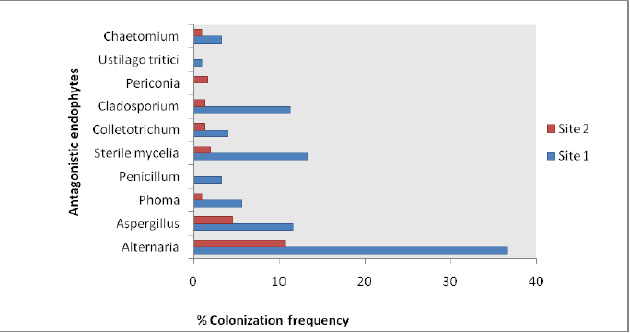 |
Figure 2: % Colonization frequency of the antagonistic endophytic population from both sites. |
Characterization of antagonistic fungal endophytes
Morphological characterization of antagonistic fungal endophytes: Based on the morphology different antagonistic endophytic fungal isolates were recovered on PDA plate. Among 40 antagonistic endophytic isolates 62.5% showed reverse pigmentation, 15% showed velvety appearance on PDA plate while others appeared spory and cottony texture. Isolates Aspergillus niger strain 89 (NAIMCC-F-03157), Aspergillus niger strain 88, Aspergillus sp. strain 37 (NAIMCC-F-03147), Aspergillus flavipes strain 63 (NAIMCC-F-03153) and Aspergillus niger strain 50 (NAIMCC-F-03151), showed dispersed growth on PDA medium.
Functional characterization of antagonistic fungal endophytes: The antagonistic fungal endophytes recovered from Stevia, were checked for their hydrolytic potential. They were screened for multiple enzyme activity on starch, pectin, lipid, carboxy methyl cellulose, xylan and skim milk. Endophytes exhibited good amylolytic, cellulolytic, proteolytic, pectinolytic activities while xylanolytic and lipolytic activities were possesed by only few isolates. The confirmation of enzymatic activity was recorded by the presence of zone of clearance around the culture. Alternaria alternata strain 76 showed maximum zone of about 15 mm on starch agar plate while 11 mm zone was recorded for cellulolytic activity by isolate Alternaria alternata strain 26.Maximum pectinolytic activity was recorded by isolate Alternaria alternata strain 99, whereas isolate Alternaria alternata strain 4 (NAIMCC-F-03138), showed maximum zone (11 mm) of clearance on CMC plate. The isolates Aspergillus sp. strain 46, Phoma sp. strain18, Phoma sp. strain 33 and Alternaria alternata strain 94, Aspergillus niger strain 50, Alternaria porri strain 6 (NAIMCC-F-03139), and Alternaria brassicae strain 5 (NAIMCC-F-03140), showed minimum inhibition (2mm) on same substrate.
Plant growth promotiong attributes of antagonistic endophytes:The antagonistic endophytes were studied for plant growth promoting traits such as siderophore production, phosphate solubilisation, Indole acetic acid production. A comprehensive overview of the PGP traits of antagonistic endophytes is given in the Table 2. About 40% of the antagonistic endophytes were positive for siderophore production and the amount of siderophore produced ranged between 1.3-27 mgml-1. On Pikovyaskya’s agar, 31% of the antagonistic endophytes showed phosphate solubilisation; efficiency of P solubilisation ranged between 5.4%-85.7percent. Based on the results isolates Phoma sp. strain 18 and Sterile mycelia strain 3 were found to be most efficient P solubilizer. A total of 29% of the antagonistic endophytes were positive for IAA production which ranged between 1.9-11µgml-1 (Table 2).
| Table 2: Plant growth promoting attributes of antagonistic endophytes | |||||
| S.no | Isolates | Identity | IAA (µg/ml) | Siderophore (mg/ml) | % solubilising efficiency of Phosphate |
| 1 | SR/II/2 | Alternaria porri | – | 8.42±0.20 | 75 |
| 2 | SR/II/4 | Alternaria alternata | 6.2±0.03 | 1.6±0.20 | 52 |
| 3 | SR/II/5 | Alternaria brassicae | – | 3.79±0.05 | 71.4 |
| 4 | SR/II/6 | Alternaria porri | 3±0.04 | 6.2±0.06 | 60 |
| 5 | SR/II/42 | Alternaria sp. | 8±0.03 | 3.72±0.08 | – |
| 6 | SR/II/45 | Penicillium mallochii | 6.3±0.05 | 4.2±0.04 | 32 |
| 7 | SR/II/50 | Aspergillus niger | 4.8±0.01 | – | 6.25 |
| 8 | SR/II/52 | Phoma sp. | – | 6.6±0.03 | 10 |
| 9 | SR/II/54 | Phoma sp. | 4.6±0.02 | 8.7±0.03 | 20 |
| 10 | SR/I/76 | Alternaria alternata | – | 3.2±0.02 | 5.4 |
| 11 | SR/I/77 | Alternaria sp. | 5.5±0.03 | 6.9±0.02 | 22 |
| 12 | SR/I/78 | Alternaria tenuissima | 6.5±0.02 | 8.02±0.03 | 11 |
| 13 | SR/I/94 | Alternaria alternata | 3.8±0.01 | 2.8±0.03 | 8 |
| 14 | SR/I/99 | Alternaria alternata | 5.3±0.3 | – | 36 |
| 15 | SR/II/63 | Aspergillus flavipes | 4.6±0.04 | 11.12±0.16 | 22.2 |
| 16 | SR/II/1 | Alternaria alternata | – | 17.09±0.1 | – |
| 17 | SR/II/9 | Alternaria alternata | 9.8±0.6 | 7.45±0.12 | 17.3 |
| 18 | SR/II/20 | Alternaria sp. | 6.3±0.06 | 8.03±0.11 | 70 |
| 19 | SR/II/21 | Alternaria alternata | – | 8.03±0.11 | 50 |
| 20 | SR/II/23 | Chaetomiumglobosum | 6.7±0.05 | 15.51±0.12 | 14 |
| 21 | SR/II/24 | Alternaria alternata | – | 14.63±0.14 | 12 |
| 22 | SR/II/26 | Alternaria alternata | 1.9±0.04 | 3.15±.10 | 10 |
| 23 | SR/II/33 | Phoma sp. | 4.5±0.01 | 1.66±0.031 | – |
| 24 | SR/II/36 | Alternaria brassicae | 8.2±0.7 | 27±20.22 | 65 |
| 25 | SR/II/55 | Alternaria alternata | – | 4.42±0.14 | 14 |
| 26 | SR/II/60 | Ustilago tritici | 5.5±0.04 | 9.51±0.17 | – |
| 27 | SR/I/72 | Alternaria brassicae | – | 23±0.08 | 34 |
| 28 | SR/II/81 | Alternaria alternata | – | 22±0.06 | – |
| 29 | SR/I/85 | Alternaria alternata | 5.7±0.05 | 6.07±0.03 | 37 |
| 30 | SR/I/95 | Alternaria alternata | – | 4.5±0.04 | – |
| 31 | SR/II/100 | Alternaria alternata | 5.8±0.07 | 5.8±0.07 | 19 |
| 32 | SR/I/83 | Alternaria alternata | 4.5±0.05 | 6.18±0.19 | – |
| 33 | SR/II/18 | Phoma sp. | – | 1.3±0.02 | 85.7 |
| 34 | SR/II/37 | Aspergillus sp. | 11±0.01 | 13.3±1.12 | – |
| 35 | SR/II/46 | Aspergillus sp. | 6.8±0.06 | 8.06±0.07 | – |
| 36 | SR/I/88 | Aspergillus niger | 8.1±0.2 | – | – |
| 37 | SR/I/89 | Aspergillus niger | 11.2±0.4 | 6.72±1.4 | – |
| 38 | SR/I/90 | Alternaria alternata | – | – | 35 |
| 39 | SR/II/3 | Sterile mycelia | – | 8.42±0.20 | 85.7 |
| 40 | SR/II/17 | Alternaria brassicae | 6.25±0.05 | 24.33±0.12 | – |
Sequence analysis of ITS region of rDNA gene fragments
All the selected isolates produced a single PCR product with approximately 600 bp. Purification of the PCR product was performed using Banglore Genei purification kit and sequencing was performed by Xcelris Genomics, Ahemdabad using the same set of primers as mentioned earlier. The full length sequences of the isolates were compared with the related fungal sequences in the GenBank databases and sequence similarities were determined using BLAST sequence similarity search tool (Altschul et al., 1990). The sequences of ITS region of rDNA gene of the fungal endophytes were deposited in the GenBank and given accession numbers (Table 3).
| Table 3: Genetic relatedness of GenBank to antagonistic fungal endophytes recovered from Stevia rebaudiana Bertoni using ITS rDNA gene sequence analysis | ||||
| Isolate No. | Site | Accession no. | % Similarity | Organism |
| SR/II/2 | 1 | KJ592050 | 99% | Alternaria porri |
| SR/II/4 | 1 | KJ592051 | 100% | Alternaria alternata |
| SR/II/5 | 1 | KJ592052 | 99% | Alternaria brassicae |
| SR/II/6 | 1 | KJ603463 | 98% | Alternaria porri |
| SR/II/42 | 1 | KJ713969 | 100% | Alternaria sp. |
| SR/II/45 | 1 | KJ713970 | 99% | Penicillium mallochii |
| SR/II/50 | 1 | KJ648618 | 100% | Aspergillus niger |
| SR/II/52 | 1 | KJ713971 | 99% | Phoma sp. |
| SR/II/54 | 1 | KJ648619 | 99% | Phoma sp. |
| SR/I/76 | 1 | KJ713972 | 99% | Alternaria alternate |
| SR/I/77 | 2 | KJ713973 | 100% | Alternaria sp. |
| SR/I/78 | 1 | KJ728832 | 100% | Alternaria tenuissima |
| SR/I/94 | 1 | KJ728833 | 100% | Alternaria alternate |
| SR/I/99 | 1 | KJ728834 | 99% | Alternaria alternate |
| SR/II/63 | 1 | KF671231 | 94% | Aspergillus flavipes |
| SR/II/1 | 1 | KJ728835 | 100% | Alternaria alternate |
| SR/II/9 | 1 | KJ735925 | 99% | Alternaria alternate |
| SR/II/20 | 1 | KJ728836 | 98% | Alternaria sp. |
| SR/II/21 | 1 | KJ728837 | 99% | Alternaria alternata |
| SR/II/23 | 1 | KJ728838 | 100% | Chaetomiumglobosum |
| SR/II/24 | 1 | KJ728839 | 99% | Alternaria alternate |
| SR/II/26 | 1 | KJ728840 | 100% | Alternaria alternate |
| SR/II/33 | 1 | KJ728841 | 100% | Phoma sp. |
| SR/II/36 | 1 | KJ728842 | 99% | Alternaria brassicae |
| SR/II/55 | 1 | KJ728843 | 99% | Alternaria alternata |
| SR/II/60 | 1 | KJ735919 | 100% | Ustilago tritici |
| SR/I/72 | 1 | KJ735920 | 99% | Alternaria brassicae |
| SR/II/81 | 1 | KJ735921 | 100% | Alternaria alternata |
| SR/I/85 | 1 | KJ735922 | 100% | Alternaria alternata |
| SR/I/95 | 1 | KJ735923 | 100% | Alternaria alternata |
| SR/II/100 | 1 | KJ735924 | 100% | Alternaria alternata |
| SR/I/83 | 2 | KJ748009 | 100% | Alternaria alternata |
| SR/II/18 | 1 | KJ748010 | 100% | Phoma sp. |
| SR/II/37 | 1 | KJ767528 | 100% | Aspergillus sp. |
| SR/II/46 | 1 | KJ767529 | 100% | Aspergillus sp. |
| SR/I/88 | 1 | KJ767530 | 100% | Aspergillus niger |
| SR/I/89 | 1 | KJ767531 | 100% | Aspergillus niger |
| SR/I/90 | 1 | KJ767532 | 100% | Alternaria alternata |
| SR/II/3 | 1 | NS | NS | Sterile mycelia |
| SR/II/17 | 1 | KJ767533 | 100% | Alternaria brassicae |
| 1= Misrod Agriculture Field 2= Green House NS= Not Sequenced | ||||
 |
Figure 3: % Dominance of the antagonistic endophytic population from both sites. |
Phylogenetic analysis
To know the phylogenetic relationship among the isolates and also to confirm their taxonomical status, certain ITS rDNA sequences were chosen from GenBank databases via BLAST search analysis. The sequences were chosen from the top 20 database hits obtained in the blast search by querying the obtained sequences individually. These sequences were aligned using CLUSTAL W 1.83 (Thompson et al., 1994). Phylogenetic trees were generated by neighbourhood joining method with 100 bootstrapping replicates using MEGA version 5.( Fig:4)
 |
Figure 4: Phylogenetic tree generated on the basis of ITS rDNAsequences of antagonistic endophytic fungal isolates aligned with sequences in GenBank databases |
Discussions
Characterization of fungal endophytes from Stevia rebaudiana Bertoni was considered important because only few attempts have been made earlier to characterize fungal endophytes from this useful plant, (Begum et al 2008; Kumari and Chandra 2013). Various fungal diseases have been reported to pose serious problems to S.rebaudiana Bertoni commonly known as Stevia, a popular non calorific sweetener. These include Verticillium dahlia on leaves (Farrar et al., 2000), S. sclerotiorum reported in Canada (Chang et al., 1997), S. rolfsii in India (Kamalakannan et al., 2007) and Botryis cinerea in Itlay (Garibaldi et al., 2009). Sclerotinia stem rot (white mold) of soybean was first reported in Hungary in 1924 and since has been reported in Argentina, Brazil, Canada, India, Nepal, South Africa and United States. A great economic loss to crop plants by different phytopathogens results in low yields. The species composition of the endophytic assemblage and frequency of infection varies according to host species and site characteristic such as elevation, exposure, associated vegetation, tissue type (Fisher et al., 1994) and tissue age (Fisher et al., 1986; Rodrigues, 1994).
In the present study, foliar endophytic microorganisms were studied using various diversity indices viz Simpson index (Simpson, 1949), Shannon index (Shannon and Weaver, 1949), evenness index (Ludwig and Reynolds, 1988) and richness index (Margalef, 1958; Menhinick, 1964). Higher value of Shannon index and evenness index with lower values of Simpson index indicated greater diversity. Bills et al. (2002) described a significant difference between tropical and temperate endophytes, in terms of their ability to produce number of bioactive natural compounds isolated from endophytes. This observation suggests the importance of the host plant in influencing the general metabolism of endophytic microbes.
Among 339 recovered endophytic fungal isolates, 40 isolates were screened out as potential antagonistic endophytes against broad spectrum plant pathogen S. sclerotiorum following using dual culture technique. It was found that maximum numbers of antagonistic endophytes were recovered from site1 as compared to site 2. This may be explained as site 1 is open agricultural field which was exposed to wide variety of phytopathogens so in order to overcome these pathogens several bioactive compounds are produced by them whereas site 2 is closed area which was exposed to limited number of phytopathogens. Sadrati et al. (2013) screened 20 endophytic fungi from wheat which showed antimicrobial activities against 12 pathogenic bacteria, yeast and two phytopathogenic fungi. Percentage growth inhibition ranged between 6.9%-19% after 24 hr of incubation. Diversity analysis of the antagonistic endophytic population showed significant diversity index values. Maximum antifungal activity against S. sclerotiorum was recorded by Aspergillus flavipes strain 63 followed by strain Aspergillus niger strain 89 and Alternaria alternata strain 99 after 24 hrs of incubation.
For instance, site1 is open agricultural field with surrounding vegetation like wheat and soybean, which favours the establishment of endophytic colonization whereas site 2 is devoid of natural open conditions of environment which seems to be the major factor for tissue specific fluctuations in the recovery of endophytes The increased species richness in foliar tissues may be a result of super infection of the leaves overtime by airborne inocula (Carroll et al., 1977; Suryanarayanan and Vijaykrishna, 2001).
The 40 antagonistic endophytic fungal isolates were studied for morphological, functional and genotypic characterization. In the present study, significant functional diversity was observed among the antagonistic endophytes with respect to their hydrolytic potential viz. amylolytic, cellulolytic, lipolytic, proteolytic, pectinolytic and xylanolytic activities. 19% of the antagonistic endophytic fungal population showed amylase, cellulase, pectinase and protease production whereas 17% were xylanase producing and rest 7% were lipase producers. Fifty fungal strains isolated from medicinal plants (Alpinia calcarata, Bixa orellana, Calophyllum inophyllum and Catharanthus roseus) showed 64% were lipase producers, 62% were amylase and pectinase whereas 32% were cellulase and 30% were laccase producers (Sunitha et al 2013). Begum et al. (2008) reported that majority of the endophytes from Stevia leaves were cellulolytic in nature. Suganthi et al. (2011) isolated and characterized Aspergillus niger (BAN 3E) out of five fungal isolates as the most potent á-amylase producer. Sidkey (2011) found endophytic strain F2Mbb to produce extracellular amylase.
These potential antagonistic endophytes were screened for plant growth promoting attributes like siderophore production, Indole acetic acid production and phosphate solubilisation. In present study, about 40% of the antagonistic endophytes were positive for siderophore production and amount of siderophore produced ranged between 1.3-27 mg ml-1, 31% of the antagonistic endophytes showed positive results for phosphate solubilisation. Phosphate solubilisation efficiency ranged between 5.4-85.7% whereas 29% of the antagonistic endophytes were recorded positive for IAA production between the range 1.9-11 µg ml-1.Certain endophytes were observed to improve the ecological adaptability of host enhancing their tolerance to environmental stress and resistance to phytopathogens (Kimmon et al., 1990; Struz et al., 1999). In a study performed on Absidia corymbifera, fungi isolated from rhizospheric soil, were found to produce siderophore in the range of 4-4.55 µg ml-1 (Holzberg and Artis, 1983). Maliha (2004) found Aspergillus flavus, Aspergillus niger and P.canescens as the most potent phosphate solubilizers, (Bilal et al, 2018).
Sequence analysis revealed that majority of fungal endophytes belonged to Alternaria alternata followed by Aspergillus niger, Phoma, Chaetomium globosum, and Ustilago tritici. Mandyam et al. (2010) employed sequencing of ITS region for studying Dark septate endophytes (DSE) in annually burned tallgrass prairie. In a nut shell, present investigation has shown that Stevia rebaudiana Bertoni harbours a good deal of antagonistic fungal endophytic community. These endophytes have exhibited various characteristics features which may pose better fitness to Stevia plant and reveal ecological significance of endophyte- host relationship.
Acknowledgements
First author is supported as Junior Research Fellowship from M.P.Biotechnology Council, Bhopal. Authors are grateful to Dr S.K. Singh, Coordinator, National Facility (NFCCI), Pune for identification of fungi. Financial support from NASI for sequencing of the samples is highly acknowledged. First author is extremely thankful to Dr. S.K. Sharma, Incharge NAIMCC at ICAR-NBAIM, Maunath Bhanjan (U.P) for culture deposition at his facility and providing accessions numbers. Authors are also thankful to Dr. Nidhi Gujar for their help in conduct of this research.
References
Altschul SF, Gish W, Miller W, Myers EW and Lipman DJ (1990). Basic local alignment search tool. J Mol Biol 215(3):403-410.
Aneja K R, Experiments in Microbiology (2003). Plant Pathology and Biotechnology, 4th edition, New Age International.
Ankita Verma, B.N.Johri and Anil Prakash (2014). Antagonistic Evaluation Of Bioactive Metabolite From Endophytic Fungus, Aspergillus flavipes KF671231. Journal of Mycology, Article ID 371218, 5 pages.
Arnold AE, Henk DA, Eels RL, Lutzoni F and Vilgalys R (2007). Diversity and phylogenetic affinities of foliar fungal endophytes in loblolly pine inferred by culturing and environmental PCR. Mycologia 99(2) 185-206.
Begum R (2010). Bioprospecting of endophytes from Stevia rebaudiana Bertoni. PhD Thesis. Barkatullah University, Bhopal.
Bernstein ME, Carroll GC (1977). Internal fungi in old growth Douglas fir foliage. Can J of Bot 55: 644-653
Bhagopaty RK, Joshi SR (2011) Multilocus Molecular Characterization of endophytic fungi isolated from five medicinal plants of Meghalaya, India. Microbiology 39 ( 2): 71-78.
Bilal L & Asaf S & Hamayun M, Gul H, Iqbal A, Ullah I, Lee IJ, Hussain A (2018). Plant growth promoting endophytic fungi Asprgillus fumigatus TS1 and Fusarium proliferatum BRL1 produce gibberellins and regulates plant endogenous hormones. Symbiosis 2: 117-127.
Bills GA, Dombrowski F, Polishhook PJ (2002). Recent and future discoveries of pharmacologically active metabolite from tropical fungi. In: R Watling, J.C.Frenkland, A.M. Aineworth, S.Issac and C.H. Robinson (ed), Tropical mycology: Micromycetes vol 2 CABI Publishing , New York, pp. 165-194
Brick JH, Bostock RM, Silverstone SE (1991). Rapid in situ assay for indole acetic acid production by bacteria immobilized on nitrocellulose membrane. Appl Environ Microbiol 57:535-538.
Chang KF, Howard RJ, Guadiel RG (1997). First report of stevia as a host for Sclerotinia sclerotiorum. Plant Dis 81(3): 311.
Choedon T , Mathan G, Arya S, Kumar VL (2006). Anticancer and cytotoxic properties of the latex of Calotropis procera in a transgenic mouse model of hepatocellular carcinoma. World J Gastroenterol.
Choi YW, Hodgkiss IJ, Hyde KD (2005). Enzyme production by endophytes of Brucea javanica . J Agricult Technol pp 55-66.
Clay K, Schardl C (2002). Evolutionary Origins and Ecological Consequences of Endophyte Symbiosis with Grasses. Am Nat 160: 99-127.
Costa JM, Loper JE (1994). Characterization of siderophore production by the biological control agent Enterobacter cloacae. Mol Plant-Microbe Interact 7: 440–448.
De Bach PH (1964). Biological control of Insects Pests and weeds. New York: Reinhold.
Dissanayake RK, Ratnaweera PB, Williams DE, Wijayarathne CD, Wijesundera RLC, Andersen RJ, de Silva ED (2016). Antimicrobial activities of mycoleptodiscin B isolated from endophytic fungus Mycoleptodiscus sp. of Calamus thwaitesii Becc. J Appl Pharm Sci. 6: 1-6.
Farrar, JJ, Davis RM (2000). First report of Verticillium dahlia On Stevia rebaudiana) in North America. Plant Dis 84: 922.
Fisher PJ and Petrini O (1987). Tissue specificity by fungi endophytic in Ulex europaeus. Syddowia 40: 46-50.
Fisher PJ Anson AE and Petrini O (1986) Fungal endophytes in Ilex gallii. T Brit Mycol Sco 86 (1): 153- 193.
Fisher PJ, Petrini O, Petrini LE, Sutton BC (1994). Fungal endophytes from the leaves and twigs of Quercus ilex L. from England, Majorca an Switzerland. New Phytologists 127, 133-137.
Garibaldi A, Bertetti D, Gullino ML (2009). First report of Botrytis blight caused by Botrytis cinerea on flowering Dogwood (Cornus florida) in Italy. J Plant Dis 93(5): 549.
Gond S, Verma V, Kumar A and Kharwar R (2007). Study of endophytic fungal community from different parts of Aegle marmelos correae (Rutaceae) from Varanasi ( India). World J Microbiol Biotechnol 23, 1371-1375.
Halmschlager E, Butin H, Donaubauer E. (1993) Endophytic fungi in leaves and twigs of Quercus petraea. European Journal of Forest Pathology 23: 51–63.
Hamzah TNT, Lee SH, Hidayat A, Terhem R, Hanum IF, Mohamed R (2018). Diversity and characterization of endophytic fungi isolated from the tropical Mangroove species, Rhizophora mucronata, and identification of potential antagonists against the soil borne fungus, Fusarium solani 9: 1-17.
Hata K, Futai K (1995). Endophytic fungi associated with healthy pine needles and needles infested by the pine needle gall midge Thecodiplosis japonensis. Can J Bot 73: 384-390.
Hawksworth DL, Kirk PM, Sutton BC, Pegler DM (1995). Ainsworth & Bisby’s dictionary of the fungi. 8th edition. CAB International, Wallingford.
Higgins KL, Arnold AE, Miadlikowska J, Sarvate SD, Lutzoni F. (2007) Phylogenetic relationships, host affinity, and geographic structure of boreal and arctic endophytes from three major plant lineages. Molecular Phylogenetics and Evolution 42: 543–555.
Holzberg M, Artis WM (1983). Hyroxamate siderophore production by opportunistic and systemic fungal pathogens. Infect Immun 40(3): 1134-1139.
Howell CR (2003). Mechanisms employed by Trichoderma species in the biological control of plant diseases: the history and evolution of current concepts. Plant Dis. 87, 4-10.
Jeewon R, Ittoo J, Mahadeb D, Jaufeerally Y, Wang HK, Liu Pong Ai ( 2013). DNA based Identification and Phylogenetic characterization of endophytic and saprobic fungi from Antidesna madagascariense, a Medicinal Plant in Mauritius. J Mycol 1-10.
Kamalakannan A,Valluvaparidasan V, Chitra K, Rajeswari E, Salaheddin K, Ladhalakshmi D and Chandrasekaran ( 2007). First report of root rot of Stevia caused by S.rolfsii in India. J of Plant Pathol 56 (2): 350.
Kimmons CA Gwinn KD and Bernard EC (1990). Nematode reproduction on endophyte infected and endo free tall fescue . Plant Dis 74, 757-761.
Kumaresan V, Suryanarayanan TS (2002). Endophyte assemblages in young, mature and senescent leaves of Rhizophora apiculata: evidence for the role of endophytes in mangrove litter degradation. Fungal Diversity 9: 81-91.
Lavini A, Riccardi M, Pulvento C, Luca SD, Scamosci M, Andria RD (2008). Yield, Quality and Water Consumption of Stevia rebaudiana Bertoni Grown under Different Irrigation Regimes in Southern Italy. Italian Journal of Agronomy 2:135-143.
Lee Sunhu, Encarnacion FM, Zentella CZ, Flores Gracia L, Escamilla JE and Kennedy C (2004). Indole acetic acid biosynthesis is deficient in Glucanacetobacter diazotrophicus strains with mutation in cytochrome C Biogenesis Genes. 186 (16): 5384-5391.
Lodge DJ, Fisher PJ and Sutton BC (1996). Endophytic fungi of Manilkara bidentata leaves in Puerto Rico Mycologia 88: 733-738.
Ludwig JA, Reynolds JF (1988). Statistical Ecology. A primer on methods and computing. John Wiley and Sons, New York, p. 337
Lumyong, S, Techa W, Lumyong P et al (2009). Endophytic fungi from Calamus kerrianus and Wallichia caryotoides (Arecaceae) at Doi Suthep- Pui National Park, Thailand. Chiang Mai Journal of Science, 36(2), 158-167.
Madhumita K and Chandra S. (2013). Localisation and isolation of fungal endophytes from healthy tissue of Stevia rebaudiana (Bert.). Internal journal of phytomedicine 5:435-440.
Maliha R, Samina K, Najma A, Sadia A, Farooq L (2004). Organic acids production and phosphate solubilisation by phosphate solubilising microorganism under in vitro conditions. Pakistan. J Biol Sci 7: 187-196.
Mandyam K, Loughin T and Jumpponen A (2010). Isolation and morphological and metabolic characterization of common of endophytes in annually burned tallgrass prairie. Mycologia, The Mycological Society of America, Lawrence 102 (4) KS 66044- 8897.
Margalef R (1958). Temporal succession and spatial heterogeneity. In: Perspectives in Marine biology, Buzzati-Traverso (ed). Univ California Press, Berkeley, pp- 323-347.
Menhinick EF (1964) A comparison of some species- individual diversity indices applied to samples of field insects. Ecology 45: 859-861.
Midmore D.J., Rank A.H. (2002). A new rural industry – Stevia – to replace imported chemical sweeteners. Report for the Rural Industries Research and Development Corporation 02/022, 55 p.
Mishra KM, Anuradha P, Rao DS (2013). Screening a Pectinolytic fungal strain; Aspergillus foetidus MTCC 10367 for the production of multiple enzymes of industrial importance. International Journal of Pharm and Bio Sciences. 2:1205 – 1209.
Newman JD, Cragg Gorden M, Snader KM (2003) Natural Products as sources of new drugs over the period 1981-2002. J Natural Prod1022-1037.
Paterson RRM, Bridge PD (1994). Biochemical Methods for Filamentous Fungi. IMI Technical Handbooks No. 1. Wallingford, UK: CAB International
Pikovskaya RI (1948). Mobilization of phosphorus in soil in connection with the vital activity of some microbial species. Microbiology 17:62-370
Pirttila A, Joensu P Pospiech H Jalonen J, Hohtola A (2004). Bud endophytes of Scots pine products adenine derivatives and other compounds that affect morphology and mitigate browning of callus cultures. Physiol Plant 121: 305-312.
Pointing SB, Buswell JA, Vrijmoed LLP, Jones EBG (1999). Extracellular cellulytic enzyme profiles of ligninolous mangrove fungi. Mycol Res 103(6): 696-700.
Prakash A, Begum R, Johri BN. (2008). Diversity of endophytic fungi in leaves of Stevia rebaudiana Bertoni. In Proceedings of the 12th International symposium on microbial ecology: 17-22 August, Crains Australia.
Ratnaweera PB, de Silva ED. Endophytic fungi: A remarkable source of biologically active secondary metabolites (2017). In: Endophytes: Crop productivity and protection. Sustainable development and biodiversity. Maheshwari D, Annapurna K. (eds). Vol 16. Springer, Cham, 2017, pp 191-212.
Ratnaweera PB, Walgama RC, Jayasundera KU, Herath SD, Abira S, Williams DE, Andersen RJ and de Silva ED (2018). Antibacterial activities of endophytic fungi isolated from six Sri Lankan plants of the family Cyperaceae. Bangladesh Journal of Pharmacology 13:264-272.
Rifai MA (1969) A revision of the genus Trichoderma. Mycological Papers.116: 1-56.
Rodriguez KF (1994). The foliar fungal endophytes of the Amazonian plant Euterpe Xylariaceae. Boletim do Museu Paraense Emfllio Goeldi Botanica 2 (7): 429-439.
Rodriguez KF (1994). The foliar fungal endophytes of the Amazonian plant Euterpe Xylariaceae. Boletim do Museu Paraense Emfllio Goeldi Botanica 2 (7): 429-439.
Rollinger JL and Langenhein (1993). Geographic survey of fungal endophytes community composition in leaves of coastal redwood. Mycologia 85 (2):149-153.
Sadrati N, Daoud H, Zerroug A, Dahamna S, Bounarati S (2013). Screening of antimicrobial and antioxidant secondary metabolites from endophytic fungi isolated from wheat ( T. Durum) J Plant Protection Research. 53, 128-136.
Sambrook and Russell (2001). Molecular Cloning : A laboratory Manual. Cold Spring Harbor Laboratory. 3 edition.
Schippers B, Bakker AW, Bakker PAHM (1987) Interactions of deleterious and beneficial rhizosphere microorganisms and the effect of cropping practices. Annu Rev Phytopathol. 25: 339-358.
Schulz B, Boyle C, Draenger S, Rommert AK and Krohn (2002) Endophytic fungi : a source of biologically active metabolites. Mycological Res 106: 996-11004
Schwyn B, Neilands JB (1987) Universal chemical assay for the detection and determination of siderophores. J Analytical Biochem 160: 47-56.
Sen S, Biswas G, Basu SK, Acharya K (2012). Management of leaf spot disease of Stevia rebaudiana Bertoni with antagonistic bacteria. Australian Journal of Crop Science 2:350-356.
Shakeri J, Howard A (2007) Proteolytic activity and antibiotic production by Trichoderma harzianum in relation to pathogenicity to insects. Foster Enz Microb Tech 40:961-968.
Shannon CE, Weaver W (1949) The Mathematical Theory of Communication. University of Illions Press, Urbana, USA.
Sidkey NM, MA Abo-Shadi, Balahmar R, Sabry R Badrany (2011) Purification and characterization of á amylase from newly isolated Aspergillus flavus F2Mbb.Int J of Microbiology.2,96-103.
Sierra G (1957). A simple method for the detection of lipolytic activity of microorganisms and some observations on the influence of the contact between cells and fatty substrates. Ant Van Leewenhoek 23(1): 15-22.
Simpson EH (1949). The Measurement of diversity. Nature 163-688.
Strobel G, Daisy B (2003). Bioprospecting for microbial endophytes and their natural products. Microbiology and Molecular Biology Reviews. 67:491-502.
Struz AV Christie BR, Matheson BG, Arsenault WJ, Buchanan N (1999). Endophytic bacterial community in the periderm of potato tubers and their potential to improve resistance to soil borne plant pathogens. Plant Pathol 48 : 360-370.
Suganthi R, Benazir JF, Santhi R, Ramesh Kumar V, Hari A, Meenakshi N, Nidhiya KA, Kavitha G, Lakshmi R ( 2011). Amylase production by Aspergillus niger under solid state fermentation using agro industries wastes. Inter J Eng Science and Tech.3, 1756-1763.
Sullivan O, Gara FO (1992). Traits of fluorescence Pseudomons spp. involved in suppression of plants root pathogens. Microbiol Rev 56 ( 4): 662-676.
Sunitha VH, Nirmala Devi D, Srinivas C (2013). Extracellular enzymatic activity of endophytic fungal strains isolated from medicinal plants. World J of Agicultural Sci 9(1): 01-09.
Suryanaranan TS, Thirunavukkarasu N, Govindarajulu MB , Sasse F, Jansen R, Murali TS (2009). Fungal endophytes and biosprospecting. Fungal Biol Rev 1, 11.
Suryanarayanan TS, Murali TS and Venkatesan G (2002). Occurrence and distribution of fungal endophytes in tropical forests across a rainfall gradient. Can J Bot 80: 818-826.
Suryanarayanan TS, Murali TS and Venkatesan G (2002). Occurence and distribution of fungal endophytes in tropical forests across a rainfall gradient. Can J Bot 80: 818-826.
Suryanarayanan TS, Venkatesan G, Murali TS (2003). Endophytic fungal diversity and distribution patterns. Curr Sci, 85: 489-493.
Suryanarayanan, TS and Vijaykrishna D (2001). Fungal endophytes of aerial roots of Ficus benghalensis. Fungal Diversity, 8; 155-161.
Szekeres A, Leitgeb B, Kredics L, Antal Z, Hatvani L, Manczinger L, Vagvolgyi C (2005) Peptaibols and related peptaibiotics of Trichoderma—a review. Acta Microbiol Immunol Hung 52:137–168.
Taylor, 1.E., Hyde, K.D. and Jones, E.B.G. (1999). Endophytic fungi associated with the temperate palm, Trachycarpus fortunei, within and outside its natural geographic range. New Phytologist 142: 335-346.
Teather RM, Wood PJ (1982). Use of Congo red polysaccharide interactions in enumeration and characterization of cellulolytic bacteria from bovine rumen. Appl Environ Microbiol 43 (4):777-780.
Thompson JD, Higgins DG, Gibson TJ (1994). Clustal W: Improving the sensitivity of progressive multiple sequence alignment through sequence alignment through sequence weighting position- specific gap penalties and weight matrix choice. Nucleic Acids Res 22(22): 4673-4680.
Toofanee, SB and Dulymamode, R. (2002). Fungal endophytes associated with Cordemoya integrifolia. Fungal Diversity 11: 169-175.
Wakelin SA, Rosemary A, Warren Paul R, Ryder MH (2004). Phosphate solubilisation by Penicillium spp. closely associated with wheat roots. Biol Fertility Soils. 40:36-43.
Waller F, Achatz B, Baltruscha J, Fodor, Becker, K (2005) The endophytic fungus Piriformospora indica reprograms barley to salt stress tolerance, disease resistance and higher yield. Proc.Nat.Acad.Sci.USA,102:13386-13391.
White TJ, Bruns T, Lee S, Taylor JW (1990). Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. In: PCR Protocols: A Guide to Methods and Applications, (eds. Innis MA, Gelfand DH, Sninsky JJ, White TJ) Academic Press, Inc., New York. pp. 315-
322.
Wilson D, Carroll (1994). Infection studies of Discula quercina, an endophyte of Quercus garryona. Mycologia 86 (5): 633-
647.


