1Applied Biotechnology Research Centre, Baqiyatallah University of Medical Sciences, Tehran, Iran,
2Biology Research Centre, Faculty of Basic Sciences, Imam Hossein University, Tehran, Iran,
3Applied Microbiology Research Centre, Baqiyatallah University of Medical Sciences, Tehran, Iran,
4Department of Industrial and Environmental Biotechnology, National Institute of Genetic Engineering and
Biotechnology, Tehran, Iran
5Plant biotechnology Department, National Institute for Genetic Engineering and Biotechnology (NIGEB), Tehran, Iran,
Article Publishing History
Received: 31/05/2016
Accepted After Revision: 25/06/2016
Astonishing improvements in information technology, in combination with the valuable experimental data gathered during the past decades, has revolutionized vaccine design strategies. On the other hand, the development of genetic engineering methods has enabled us to design and produce new chimeric proteins for many different purposes, including immunization. In this study we took the advantages of these improvements to develop an efficacious vaccine against three important enteropathogenic bacteria: enterotoixigenic E. coli (ETEC), enterohemorragic E. coli (EHEC), and Shigella. To do this, appropriate immunogens, including subunit B of heat labile toxin and mutated STa toxin from ETEC; EspA and Stx B from EHEC; and the N-terminal part of IpaD, an immunogen from diffrent Shigella strains, were selected. These proteins were fused together by an appropriate peptide linker. Bioinformatic analyses, including the physicochemical parameters calculation, secondary and tertiary structures prediction and verification, antigenic B-cell and T-cell epitopes prediction were performed. Then the protein was reverse-translated to DNA and was codon optimized for expression in E. coli cells. The results showed a proper 3D structure, good theoretical immunogenicity, invitro and invivo stability of the chimeric designed protein and proper structure and stability of the related mRNA. Altogether, the results suggest that the designed chimeric protein, structurally and immunologically, has almost all factors of an efficient vaccine candidate and can be tested in experimental studies.
Chimeric Antigen, In Silico Analysis, Candidate Vaccine Design, Etec, Ehec, Shigella
Hajizade A, Ebrahimi F, Amani J, Arpanaei A, Salmanian A. H. Design and in Silico Analysis of Pentavalent Chimeric Antigen Against Three Enteropathogenic Bacteria: Enterotoxigenic E. Coli, Enterohemorragic E. Coli And Shigella. Biosc.Biotech.Res.Comm. 2016;9(2).
Hajizade A, Ebrahimi F, Amani J, Arpanaei A, Salmanian A. H. Design and in Silico Analysis of Pentavalent Chimeric Antigen Against Three Enteropathogenic Bacteria: Enterotoxigenic E. Coli, Enterohemorragic E. Coli And Shigella. Biosc.Biotech.Res.Comm. 2016;9(2). Available from: https://bit.ly/2BegLR1
Introduction
Information technology has revolutionized almost all fields of sciences and technologies, including vaccinology and vaccine development strategies (Lund 2005). There are many authentic software and programs that could serve as powerful tools for rational designe of new vaccines. There are such tools in almost all steps of a vaccine design project, from determination of a hypothetical immunogen structure (Pollastri and Mclysaght 2005, Yang, Yan et al. 2015) to the evaluation of its interaction with cells and molecules of immune system (Saha and Raghava 2004, EL‐Manzalawy, Dobbs et al. 2008). By applying these tools, the time and costs of a vaccine development project will be dramatically reduced. Indeed, we proceed through a project clearly, and as a result, we are able to manipulate the project more efficiently. Having hypothetical antigens of a pathogen, it is possible to evaluate the effectiveness of these antigens as candidate vaccines and choose the most potent ones for in vivo analysis, (Davies and Flower 2007 and Sharma, et al. 2016).
By doing more researches and gathering more experimental data, in combination with development of new algorithms and software, the science of computational vaccinology will be developed, so that it is not out of reach to stand on a point where we have the ability to design a multivalent efficacious immunogen (against several pathogens) (Pinheiro, Martins et al. 2011). The process, despite being new, is already used in an increasing manner for evaluating the effectiveness of different antigens or proposed chimeric antigens as candidate vaccines.
On the other hand, the development of genetic engineering methods has enabled us to design new chimeric proteins that carry more than one epitope from one or more pathogen, as a candidate vaccine against these pathogens. Chimeric antigens could be designed to carry different epitopes, so it is postulated that they can elicit immune responses against all included antigens (Ahlers et al. 2001). A striking advantage of these vaccines is the fact that “working on one protein (i.e. chimeric protein) is preferable than working on several proteins”; it reduces the cost and time greatly.
Annually, there are more than 800,000 deaths from diarrheal diseases, more than AIDS, malaria, and measles combined (Liu, Johnson et al. 2012). The diseases accounts for 1 in 9 child deaths worldwide and are the second leading cause of death in children under 5. Enteropathogenic bacteria are of the main causative agents of diarrhea. Shigella species, ETEC, and EHEC are among the most important bacteria that cause the disease.
Enteropathogenic bacteria cause diarrhea all over the world. Despite all improvements in hygene standards, the problem is still remained. Indeed, phenomena like national disasters, including earthquickes and floods, create a situation that the disease can cause severe health problems. This suggests that there is a great need for developing efficacious vaccines against these pathogens. In the present study, bioinformatics tools were used for designing a chimeric antigen as a candidate vaccine against three enteropathogenic bacteria: enterotoixigenic E. coli (ETEC), enterohemorragic E. coli (EHEC), and Shigella. ETEC is a major cause of diarrhea in children under 5 in developing world and in travelers to these areas (Svennerholm and Tobias 2008, Taxt, 2016). EHEC, the pathogenic subgroup of Shiga toxin (Stx)–producing Escherichia coli (STEC), is the major etiological agent of hemorrhagic colitis and the life-threatening hemolytic uremic syndrome (HUS) (Paton and Paton 1998). Shigella species, including Shigella dysenteriae, Shigella flexneri, Shigella boydii, and Shigella sonnei, cause shigellosis, which is transmitted through faecal-oral rout and for this, it has remained a health problem, mainly in developing countries with poor hygiene standards (Hale 1991 and Brown 2016).
There have been many designated chimeric proteins against each of mentioned pathogens. To our best knowledge, it is the first report on a chimeric vaccine that may protects against these three pathogens. In this study, we designed a chimeric protein consisting of five different antigens (LTB and STa from ETEC, N-terminal part of IpaD from Shigella, and StxB and EspA from EHEC) and analyzed it by bioinformatic tools. After verifying the protein’s efficiency as a candidate vaccine, it was reverse-translated to DNA. The multigenes DNA was then codon optimized for high expression in a prokaryotic host, E. coli cells.
Methods
Immunogen Selection And Sequence Retrieval
Through the literature search and review, appropriate immunogens for each pathogen were selected. Multiple alignment softwares were used to select more prevalent and consensus sequences. Sequences and structures of the desired immunogens were adopted from UniProt (www.uniprot.org).
Construction Of Design
The order of the selected sequences was optimized for obtaining the best 3D structure and immunogenic properties. Having closer secondary and tertiary structures to the individual proteins, and also a high antigenicity of the final structure were our priorities in lining up the antigens in the chimeric structure. For a good separation, the rigid helical linker (EAAAK) with different repeats were tested to obtain a good separation of the selected antigenic domains.
Prediction Of The Protein’s Secondary Structure
Several online programs, including GOR IV and V methods (http://npsa-pbil.ibcp.fr/cgi-bin/secpredgor.pl), (Garnier, Gibrat et al. 1996), the PredictProtein Program (https://www.predictprotein.org/), Proter server (http://distill.ucd.ie/porter/) (Pollastri and Mclysaght 2005), and PSIPRED protein structure prediction server (http://globin.bio.warwick.ac.uk/psipred/ ) were used to predict the secondary structures of hypothetical proteins.
Tertiary Structure Prediction, Validation, And Refinement Of The Chimeric Protein
Online I-TASSER (iterative threading assembly refinement) software, which is based on multiple-threading alignments by LOMETS and iterative TASSER simulations(Yang, Yan et al. 2015), was used for the prediction of the tertiary structure of the chimeric protein. Raptorx (http://raptorx.uchicago.edu/) (Källberg, Wang et al. 2012), and ESyPred3D (http://www.unamur.be/sciences/biologie/urbm/bioinfo/esypred/) were also used for 3D structure prediction. Rasmol software (http://rasmol.org/OpenRasMol.html) and Accelrys Discovery Studio were used for the analysis and visualization of the structures.
All predicted protein structures have some errors (Hooft, Vriend et al. 1996), so it’s inevitable to validate the predicted models. To overcome to this problem, the best 3D model, in which the different domains were separated and exposed and had a significant score, was selected and processed by ProSA (https://prosa.services.came.sbg.ac.at/prosa) (Wiederstein and Sippl 2007) program. Indeed, via Rampage, Ramachandran diagram was plotted to determine the overall stereo-chemical quality of the model (http://mordred.bioc.cam.ac.uk/*rapper/rampage.php) (Lovell, Davis et al.).
Analysis Of The Physico-Chemical Parameters
All physico-chemical parameters, including molecular weight, theoretical isoelectric point (pI), half-life of the recombinant protein in three different hosts (E. coli, yeast, and mammalian cells), total number of positive and negative residues, extinction coefficient, instability index, and aliphatic index was calculated by the Expasy’s ProtParam tool at http://us.expasy.org/tools/ protparam.html.
Prediction Of Antigenic B- And T-Cell Epitopes
BCPred analysis tool (www.imtech.res.in/raghava/bcepred/) was used for the prediction of linear B-cell epitopes (EL‐Manzalawy, Dobbs et al. 2008). This method predicts B-cell epitopes using any of the physico-chemical properties (hydrophilicity, flexibility/mobility, accessibility, polarity, exposed surface and turns) or combination of properties (Saha and Raghava 2004). For determining conformational B-cell epitopes, Discotope (http://www.cbs.dtu.dk/services/DiscoTope/), that predicts conformational B-cell epitopes in an antigen from its amino acid sequence (Haste Andersen, Nielsen et al. 2006), and ElliPro (http://tools.immuneepitope.org/ellipro/), that uses both the sequence and three-dimensional structural data for conformational B-call epitopes (Ansari and Raghava 2010), servers were exploited. VaxiJen (http://www.ddg-pharmfac.net/vaxijen/VaxiJen/VaxiJen.html), that classifies antigens solely based on the physicochemical properties of proteins without recourse to sequence alignment (Doytchinova and Flower 2007), was also exploited, for further analyzes. For prediction of MHC class-I and class-II binding regions in the chimeric proteins, SYFPEITHI (http://www.syfpeithi.de/) (Rammensee, Bachmann et al. 1999) and ProPred (http://www.imtech.res.in/raghava/propred/) servers were used, respectively.
Codon Optimization And Mrna Structure Prediction
After confirming the efficacy of the chimeric protein as a candidate vaccine, the protein sequence was back-translated to DNA by backtranseq tool (http://www.ebi.ac.uk/Tools/st/emboss_backtranseq/). The resulted DNA was then codon-optimized by OPTIMIZER (http://genomes.urv.es/OPTIMIZER/) (Puigbo, Guzman et al. 2007). The codon-optimized sequence was further analyzed by GenScript’s patented OptimumGeneTM algorithm
For efficient translation, the final structure of the mRNA should be at the right form and energy level. Unavailable ribosome binding site (RBS) and some unfavorable secondary structures, such as long hair-pin loops have a negative impact on the translation efficiency or even could lead to translation repression. Determination of the secondary structure of the mRNA can give an insight to these problems. Here the secondary structure of the mRNA was predicted by mfold web server (Zuker 2003) at http://mfold.rna.albany.edu/?q=mfold.
RESULTS
Immunogen Selection And Sequence Retrieval
By the literature review and according to the previous studies and investigations, StxB and EspA from EHEC, LTB and modified form of STa from ETEC, and N-terminal segment of IpaD for Shigella were selected as appropriate antigens for incorporating into the chimeric protein. It has been shown that all selected antigens are potent immunogens. The amino acid sequences for Stx2B (Q7DJJ2), EspA (O33976), IpaD (P18013), STa (P01560), and LTB (P32890 ) were adopted from UniProt. The unnecessary regions of each antigen were removed so that the antigenicity remained unaffected.
Synthetic Construct Design
The potential orders of the antigens in the final structure were evaluated by two different methods: the overall antigenicity of the final structure and by comparing the similarity between secondary and tertiary structures of the each antigen alone and in the chimeric form. Moreover, placing the antigens of the same pathogen near together was important. The overall antigenicity of the various combinations of fragments was calculated by VaxiJen. Then, the ones of low antigenicity were removed and those of high antigenicity (higher than 7.2, when the threshold was 0.4) were selected for further analysis. The selected combinations were compared according to the similarity of secondary and tertiary structures’ of the individual antigens when they are alone and when they are incorporated in the chimeric antigen. The results show that the best results are met when the order of antigens is as follow: StxB-EspA-IpaD-STa-LTB (for abbreviation: SEISL), which is represented in figure 1. Having this order of antigens, the overall antigenicity of the chimeric antigen were 0.7389 and 0.847915 when analyzed by VaxiJen and ANTIGENpro (http://scratch.proteomics.ics.uci.edu/), respectively.
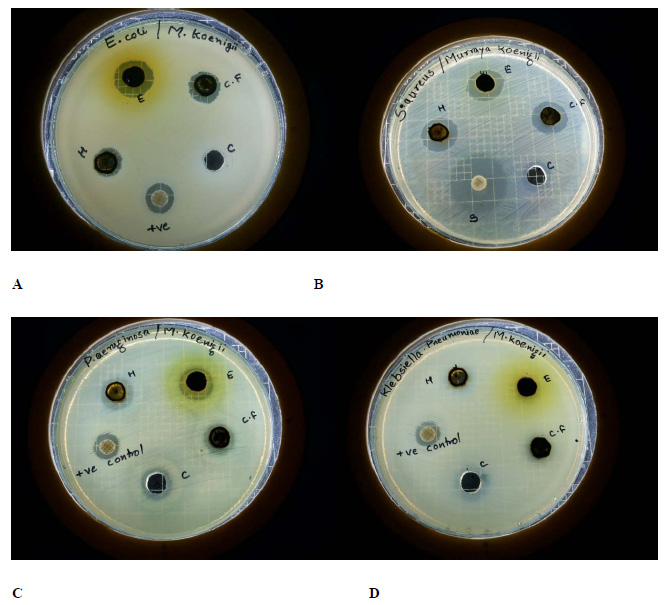 |
Figure 1: The order of the antigens in the chimeric protein. |
For effective separation between domains, an alpha helix-forming linker with the sequence of A(EAAAK)2A, which is rigid and stable, was applied (Arai, Ueda et al. 2001). A histidine tag (6xHis) was applied for facilitating the protein purification by affinity chromatography and also for the ease of detection by immunological methods.
Prediction Of The Protein’s Secondary Structure
Several methods were exploited for the prediction of secondary structure of the chimeric protein. As a control, the secondary structures of all individual antigens were either retrieved from PDB database (in the case of LTB and StxB) or predicted by different methods (all antigens). The frequency of the major types of secondary structure (alpha helix, extended strands, and random coils) in each individual antigen was compared with these structures in the chimeric antigen (Table 1). As it has been shown in table 1, different programs present different values.
| Table 1: Predicted secondary structures of chimeric protein and each fragment by different programs. | ||||||||
| Protein | Type of secondary structure | Prediction method | Native protein | Psipred | PORTER | GORIV | GORV | SSpro |
| SEISL | Alpha helix
Extended strand Random coil |
nd*
nd nd |
69
10 21 |
60
18 22 |
56
12 32 |
55
10 35 |
52
12 36 |
|
| StxB | Alpha helix
Extended strand Random coil |
15
48 38 |
10
45 45 |
16
36 48 |
14
42 44 |
10
37 57 |
11
25 64 |
|
| EspA | Alpha helix
Extended strand Random coil |
nd
nd nd |
61
5 34 |
67
2 31 |
51
16 33 |
55
10 35 |
53
17 30 |
|
| IpaD | Alpha helix
Extended strand Random coil |
nd
nd nd |
67
0 33 |
67
0 33 |
70
4 26 |
69
0 31 |
75
3 22 |
|
| STa | Alpha helix
Extended strand Random coil |
nd
nd nd |
57
0 43 |
35
20 45 |
27
18 55 |
32
0 68 |
30
16 54 |
|
| LTB | Alpha helix
Extended strand Random coil |
17
34 49 |
35
27 38 |
23
37 40 |
25
33 42 |
55
15 30 |
20
35 45 |
|
| *nd: not determined | ||||||||
By analyzing the predicted results we came to the conclusion that the prediction of the structures by the GORIV has been done more exact, so the method, which is an information theory-based and Bayesian method, was selected for the prediction of the secondary structure of the chimeric protein. Figure 2 represents the predicted secondary structure of the chimeric protein.
 |
Figure 2: Shematic view of the results for secondary structure prediction of the chimeric protein. Blue color stands for helices, purple stands for random coils, and red stands for extended strands. |
According to GOR IV, 56.47% of the secondary structure types are alpha helix, 12.02% are extended strands, and 31.51% are random coiled. PredictProtein results show that the solvent accessibility composition (core/surface ratio) for the chimeric protein was estimated to be acceptable: 60.79% of the residues were exposed with more than 16% of their surfaces.
Tertiary Structure Prediction And The Model Validation And Refinement
The tertiary structure prediction was performed by Raptorx, DISTILL, and I-TASSER methods. The accuracy of the predicted models was assessed by ModFOLD model quality assessment server (version 4.0) (http://www.reading.ac.uk/bioinf/ModFOLD/ModFOLD_form_4_0.html). The Raptorx, DISTILL, and I-TASSER methods presented models with P-values of 0.0855, 0.0915, and 0.008, respectively. The higher P-values of the first two models shows that the low confidence of the predicted models, while in the case of the predicted model by I-TASSER, the P-value is low, which it show the high confidence of the model (the P-value <0.01 means the model’s confidence is high). Figure 3 represents the predicted tertiary structure of the chimeric antigen by I-TASSER. As it can be seen, all individual domains were separated efficiently by the peptide linker, A(EAAAK)2A.
Further analysis of the predicted 3D model was carried out by ProSA-web. Providing an easy-to-use interface to the program ProSA, ProSA-web is frequently employed in protein structure validation. As it can be seen in Figure 4, the overall quality score for the model is in a range characteristic for native proteins, so the predicted structure is probably reliable.
 |
Figure 4: Tertiary protein validation by ProSA program |
Ramachandran plot analysis of the I-TASSER model (figure 5) revealed that 95.7%of the residues are in favored or allowed regions ( 88.5 % in favored region and 7.2% in allowed region) and just 4.3% of the residues are in outlier region.
Physico-Chemical Parameters
The physico-chemical parameters of the chimeric protein, calculated by Expasy’s ProtParam tool, were as the table 2. The calculation indicates that the recombinant protein is a ~ 61 kDa protein with an isoelectric point of 9.47, which shows that the protein is positively charged in physiological condition. The results also show that the expressed protein will be intact at least for 10 hours in different expression hosts. The protein’s instability index is 31.55, which indicates the protein is stable. The aliphatic index, which represents the frequency of alpha helix in a protein, was calculated as 71.74 for the chimeric protein. Higher aliphatic index indicates that the protein is more thermostable (Atsushi 1980).
| Table 2: Physicochemical parameters of the SEISL synthetic peptide. | |
| Parameter | Value |
| Number of amino acids | 551aa |
| Molecular weight | 60759.5 Dalton |
| Theoretical isoelectric point (pI) | 9.47 |
| Half-life of the protein in E. coli, in vivo | >10 hours |
| Half-life of the protein in yeast, in vivo | >20 hours |
| Half-life of the protein in mammalian reticulocytes, in vitro | >30 hours |
| Total number of positive residues | 97 |
| Total number of negative residues | 61 |
| Instability index | 31.55 |
| Aliphatic index | 71.74 |
Prediction Of Antigenic B- And T-Cell Epitopes
Prediction of continuous B-cell epitopes
The continuous B-cell epitopes were predicted by Bcepred method. The threshold for hydrophilicity, flexibility, accessibility, turns, exposed surface, polarity, and antegenic propensity were 2, 1.9, 2, 1.9, 2.4, 2.3, and 1.8, respectively. The combined threshold was calculated as 1.8, which could be considered as a good antigen for B-cell. The predicted epitopes are presented in table 2.
| Table 2: Continuous B-cell predicted epitopes by bcpred method. Based on different parameters, there are many experimentally confirmed epitopes. See the text for details. | |
| Prediction Parameter | Predicted Epitopes |
| Hydrophilicity | ADCAKGK, SKYNENDTFT, KSSTCESGSG, NNDAEAAAK, AKANEASKASTTAQK, ADVQSSTDKNAKAK, INDPRNDIS, SARSDVQS, DVNKSAQ, SAPKEAE, AKAKKKKKKKKKKNSSNYC, YKKKKKKKKKKAEA, AKAKKKKKKKKKKNSSNYC, YKKKKKKKKKKAEA, SEYRNTQ, TETKIDK, NNKTPNS |
| Flexibility | VTIKSSTCESGS, NEASKAS, ADVQSSTDKN, DYINDPR, NLLTSARSDV, QALKKDLS, YPINKDA, EAAAKAKKKKKKKKKKNSS, TGCYKKKKKKKKKK, EAAAKAKKKKKKKKKKNSS, TGCYKKKKKKKKKK, QHIDSQK, CVWNNKTP |
| Accessibility | KIEFSKYNENDTFT, KVAGKEYWTSRWNLQP, EVQFNNDAE, KANEASKASTTAQKMAN, VQSSTDKNAKAKLPQD, DYINDPRNDIS, KANNLTT, EIQQMSN, VQSLQYRT, KAIRPTNQALKKDLSQKTLTKTSLEE, DVNKSAQ, DILSNKEYPINKDARELLHSAPKEAELDG, SHRELWDKI, INNINEQYLKVYE, AAAKAKKKKKKKKKKNSSNYC, TGCYKKKKKKKKKKAEAA, AAAKAKKKKKKKKKKNSSNYC, TGCYKKKKKKKKKKAEAA, CSEYRNTQIYT, ESMAGKRE, QHIDSQKKAIERMKDTLR, AYLTETKIDKL, VWNNKTPNSI |
| Turns | YNENDTF, VQFNNDAE, TVVNNSQL, SINNINE, KKNSSNYCC, KKNSSNYCC, SMENHHHHHH |
| Exposed Surface | SKYNENDT, KANEASK, QSSTDKNAKAKLPQD, TNQALKKDLSQKTLTKT, NKEYPINKDAREL, AAKAKKKKKKKKKKNSSNY, TGCYKKKKKKKKKKAEAA, AAKAKKKKKKKKKKNSSNY, TGCYKKKKKKKKKKAEAA, HIDSQKKAIERMKDTLR, TETKIDKL, NNKTPNS |
| Polarity | KIEFSKYNE, KEYWTSR, KANEASK, DKNAKAK, KKDLSQK, TKTSLEEIALHS, KEYPINKDARELLHSAPKEAELDGYEMISHRELWDKI, YLKVYEHAV, AAKAKKKKKKKKKKNSSN, TGCYKKKKKKKKKKAEAA, AAKAKKKKKKKKKKNSSN, TGCYKKKKKKKKKKAEAA, ELCSEYR, ESMAGKREMVII, HIDSQKKAIERMKDTLR, TETKIDKL, ISMENHHHHHH |
| Antegenic Propensity | NLQPLLQS, KLPQDVI, DVQSLQYR, QLLDILS, EQYLKVYEH, SSNYCCELCCNPQCTGCYKK, SSNYCCELCCNPQCTGCYKK, VIITFKS, TFQVEVPGSQHI, KIDKLCVW |
Prediction of discontinuous B-cell epitopes
Prediction by DiscoTope server, that predicts discontinuous B cell epitopes from protein three dimensional structures, showed that there are many potent conformational B-cell epitopes all over the protein (Fig. 5). As it can be seen, each individual antigen in the final construct has conformational B-cell epitopes.
Indeed, the prediction of conformational epitopes was done by ElliPro. ElliPro, which is based on the geometrical properties of protein structure, allows the prediction of epitopes in a given protein structure or sequence. Table 3 represents the results of conformational epitope prediction by ElliPro.
Prediction of T-cell epitopes
SYFPEITHI, a database compromising more than 7000 peptide sequences known to bind class-I and class-II MHC molecules (Rammensee, Bachmann et al. 1999), was exploited for the prediction of T-cell epitopes according to HLAs corresponding to MHC class-I. Table 4 represents the scorer positions higher than 18. As it can be seen there are many epitopes that distributed along the protein sequence.
For the prediction of T-cell epitopes according to HLAs corresponding to MHC class-II, ProPred server was used. The server predicts MHC class-II binding regions in an antigen sequence, using quantitative matrices derived from published literature by Sturniolo et al. (Sturniolo, Bono et al. 1999). Analysis showed that there are many potent MHC class-II binding regions in the protein sequence (data not shown).
Codon Optimization And Mrna Structure Prediction
The reverse-translated sequence was optimized for maximal protein expression in E. coli by OPTIMIZER program and the optimized sequence was further analyzed by GenScript’s patented OptimumGeneTM algorithm. For this purpose, three parameters, including Codon Adaptation Index (CAI), average GC content, and codon frequency distribution (CFD) of the optimized sequence were analyzed. By using the algorithm the sequence’s CAI was upgraded to 0.84 (figure 6A). Possibility of high protein expression level is correlated to the value of CAI (a CAI of >0.8 is rated as good for expression in E. coli expression system). Average GC content was also reached to 47.76. Since the ideal percentage range of GC content is between 30% to 70% and there are not any peaks outside of these ranges (figure 6B), it is expected that the transcriptional and translational efficiency won’t be affected. Rare codon analysis was carried out by codon frequency distribution (CFD) graph plotting. By this, the quantities of rare codons present in the sequence were identified (figure 6C). The value of CFD for the codon with the highest usage frequency for a given amino acid in the desired expression organism is set 100. The CFD value of less than 30 is determined as low-frequency codon, which is likely to affect the expression efficiency. As it can be seen in the graph (figure 6C), there are not any codons with a CFD value of less than 30.
 |
Figure 6: Conformational B-cell epitopes of SEISL, predicted by DiscoTope server (the threshold value was set at -7.7). |
 |
Figure 6a |
 |
Figure 6b |
Prediction of mRNA secondary structure was carried out by mfold server to analyze both stability and the status of ribosome binding site (RBS). The predicted structure showed that the mRNA is stable enough to be expressed in E. coli (ΔG = −418.8 kcal/mol) and the ribosome binding site is accessible for translational machinary.
Discussion
Annually, there are more than 800,000 deaths from diarrheal diseases, more than AIDS, malaria, and measles combined (Liu, Johnson et al. 2012). The diseases accounts for 1 in 9 child deaths worldwide and are the second leading cause of death in children under five. The disease is mainly caused by microbial pathogens, although malnutrition and some illnesses can cause GI, too (Rodríguez, Cervantes et al. 2011, Oriá, et al. 2016).
Parasitic, viral and bacterial pathogens can cause the disease. Although viruses are the main diarrhagenic agents, however, enteropathogenic bacteria are very important, especially in regions with low hygiene standards. Diarrhagenic E. coli, Shigella species, Campilobacer jejuni, Vibrio cholera, and Bacterioides fragilis are the main bacterial causative of diarrhea in humans (Guarner, Khan et al. 2012). Shigella species, ETEC, and EHEC are among the most important bacteria that cause the disease. Vaccination is a good strategy for fighting against the diseases. There are many efficient candidate vaccines against each mentioned pathogens, however, there hasn’t been any vaccine that can protect against all three pathogens. Chimeric antigens, which are a combination of several antigens or antigen epitopes, have been proved to be efficient against several bacterial diseases. By having different antigens in one construct, the downstream processes, and consequently, the production time and costs will be largely diminished. Here we chose this strategy for designing of a vaccine, which simultaneously can immunize against three pathogens. The most important step in chimeric vaccine design is the selection of appropriate immunogens.
For this, we tried to choose the most immunodominant immunogen(s) of each pathogen for incorporating to the final antigen. In the case of ETEC, since pathogenic strains of the bacteria produce at least one of these two toxins, heat labile (LT) and/or heat stable (STa) (Qadri, Das et al. 2000), both toxins were selected for induction an immunity response against all pathogenic ETEC strains. LT is a member of AB5 family toxins(Sixma, Pronk et al. 1991, Sixma, Kalk et al. 1993). While the A subunit has the enzymatic activity alone, the B subunit is not poisonous lonely, however, it is able to efficiently stimulate the immune system [14]. Stable toxin type a (STa), which is a single-peptide toxin, activates guanylate cyclase resulting in ion secretion and diarrhea(Lucas 2001). Because of the low molecular weight, STa is poorly immunogenic alone and for induction of an efficient immune response, it should be coupled with a carrier protein (Zhang, Zhang et al. 2010).
It has been shown that a point mutation in the catalytic site of the toxin will diminish the toxicity of the molecule, however the mutated toxin has the ability to induce the immune responses (Taxt, Aasland et al. 2010, Zeinalzadeh, Salmanian et al. 2013). Having these findings, we chose the B subunit of LT and a mutant form of STa for immunization against all pathogenic ETEC strains. In the case of EHEC, as mentioned earlier, different combinations of EHEC virolence factors can be chosen for incorporating into a chimeric antigen. We selected the C-terminal of EspA protein and the B subunit of Stx2. EspA, a component of the pathogenicity island (Locus of Eterocyte and Effcement, LEE), is needed for the assembly of the type III secretion system (T3SS). It has been shown that in naturally infected people, the immune response against this protein is increased, so we could consider the protein as an excellent immunogen. Indeed, there are many studies that show the protectivity of this antigen in animal models (La Ragione, Patel et al. 2006, Amani, Salmanian et al. 2010). The production of shiga-like toxins is another major virolence factor of the pathogen and is associated with life-threatening hemolytic uremic syndrome (HUS), so rasing antibody against the toxins is of a great importance. There are at least two kinds of Stx that are produced by EHEC: Stx1 and Stx2; of them Stx2 is associated with more severe disease in humans (Boerlin, McEwen et al. 1999). Like LT of ETEC, the toxin is a member of AB5 family toxins and its B subunit is not poisonous alone, however, it could stimulate the human immune responses (Li, Frey et al. 2000). In the case of shigella, Ipa (insertion plasmid antigen) proteins, which are found in all Shigella species and are required for the entry of the pathogen into the host cells, are a good choice. Induction of mutation into Ipa genes cause the pathogen to lose the capability of invading host cells (Ménard, Sansonetti et al. 1993), so targeting these proteins can prevent the pathogen to invade cells. There are four major Ipa proteins: IpaA, IpaB, IpaC, and IpaD. There have been many vaccine formulations that have used a combination of these proteins. Here, the N-terminal of IpaD protein, which is immnogenicty has been proved, was selected (Hesaraki, Saadati et al. 2011).
The selection of an appropriate peptide linker could act as a bridge and connect individual components together and play a crucial role in final structure of the chimeric antigen. An ideal linker should connect the immunogens in such a way that they become completely separated in the final folded state of the chimeric protein. Linkers with á-helix structure, can meet this aim and are the best choice for this purpose. We examined many linkers and the best results were observed when the A(EAAAK)2A sequence was chosen. This amino acid repeats could produce a rigid type linker and increase the stability and folding of the chimeric antigen (Chen, Zaro et al. 2013). The predicted structure showed that the linker has successfully separated the immnogens, so that all domains are accessible to the immune system’s components. Furthermore, this linker lacks antigenic property.
It is of a great importance that the major structural elements of each individual antigen be similar to the relevant structures in the chimeric antigen. For examining this issue, five different secondary structure prediction methods (Psipred, PORTER, GORIV, GORV, and SSpro) were exploited. By comparing the calculated data from different methods with the data retrieved from PDB, we came to the conclusion that GOR IV (Garnier-Osguthorpe-Robson) method is more accurate. GOR method, developed in the late 1970s, is based on probability parameters of experimental studies of proteins that their tertiary structures are determined by X-ray crystallography, and unlike the Chou-Fasman method, is Bayesian in its analysis (Garnier, Osguthorpe et al. 1978) and this makes the prediction results more accurate. So, the secondary structure of the chimeric antigen was predicted by GOR IV method. According to the method, the most present secondary structure type in the protein are of alpha helix type, which is in agreement to the obtained results for the predicted tertiary structure of the protein.
There are two main methods for in silico tertiary structure prediction of proteins: abinitio methods, which seek to build three-dimensional protein models “from scratch”, i.e., based on physical principles rather than (directly) on previously solved structures; and comparative protein modeling, which uses previously solved structures as starting points, or templates. There are many useful programs developed according to these strategies. Here we used the hierarchical method, I-TASSER, for this purpose. The method detects the structure templates from the Protein Data Bank and by reassembling of the structural fragments from threading templates, the structure of the whole protein is constructed, so the method benefits from comparative protein modeling (Roy, Kucukural et al. 2010). The predicted structure shows that all individual immunogens are separated successfully, an issue which is necessary for a successful elicitation of the humoral immunity (Theoretically if a domain hides inside the folded protein, it won’t be accessible to antibodies and therefore, the humoral immunity won’t be evoked). However, since the tertiary structure prediction of proteins is usually with errors, it is necessary to evaluate the predicted structures. There are many algorithms and methods to do this. The evaluation of the tertiary structure by two different methods: ProSA program and Ramachandran plot analysis, shows the I-TASSER predicted model is reliable.
The physiochemical analysis of the chimeric protein shows that the protein is a positively charged in physiological conditions and it could be expressed in different expression systems. Indeed, the results show that the protein is relatively stable.
The overall antigenicity of the chimeric antigen is the most important factor for a protein to be considered as a candidate vaccine. Being identified by humoral and cellular immunity systems’ components is an essential factor for a potent immunogen. Antigenic proteins are recognized by immune systems through antigenic determinants or epitopes. B-cell and T-cell epitopes are recognized by antibodies MHC molecules, respectively. Antibodies recognize two distinct types of epitopes: continuous (linear) and discontinuous (conformational). Many valuable programs have evolved to assess the presence of each set of epitopes in a protein. Bcepred, that predicts the presence of linear B-cell epitopes based on four physico-chemical properties: hydrophilicity, flexibility, polarity and exposed surface, was exploited to predict the existence of continuous B-cell epitopes and the results showed that the experimentally confirmed epitopes are spread through the chimeric protein. Discotope and ElliPro servers were exploited for determining discontinuous epitopes. Analysis by both servers confirmed the presence of epitopes all over the protein. SYFPEITHI was used for the prediction of T-cell epitopes. The analyses showed that the chimeric antigen has many potent epitopes that can be detected by immune cells and molecules.
Having a multi-antigen protein that has almost all factors of a candidate vaccine, the protein was reverse-translated to DNA and codon optimized by GenScript’s OptimumGeneTM algorithm according to E. coli codon preferences. The rare codons were eliminated and GC content was adjusted. Morever, repeat sequences, internal chi and ribosomal binding sites, RNA instability motif and restriction sites that may interfere with cloning, were removed. The secondary structure of the resulted mRNA showed that the molecule is stable and its ribosome binding site (RBS) is accessible for ribosomes. All and all, we came to the conclusion that the designed chimeric protein could be tested in experimental studies.
Acknowledgement
This research was carried out as a part of the Ph.D thesis of Abbas Hajizade. The authors thank Applied Biotechnology Research Centre, Baqiyatallah University of Medical Sciences, for the warm and kind support.
References
Amani, J., A. H. Salmanian, S. Rafati and S. L. Mousavi (2010). Immunogenic properties of chimeric protein from espA, eae and tir genes of Escherichia coli O157: H7 Vaccine Vol. 28 No 42: Pages 6923-6929.
Ansari, H. R. and G. Raghava (2010). Identification of conformational B-cell Epitopes in an antigen from its primary sequence Immunome Res Vol.6 No 6: Pages 1-9.
Arai, R., H. Ueda, A. Kitayama, N. Kamiya and T. Nagamune (2001). Design of the linkers which effectively separate domains of a bifunctional fusion protein. Protein engineering Vol. 14 No 8: Pages 529-532.
Atsushi, I. (1980). Thermostability and aliphatic index of globular proteins. Journal of Biochemistry Vol. 88 No 6: Pages 1895-1898.
Berzofsky, J. A., J. D. Ahlers and I. M. Belyakov (2001). Strategies for designing and optimizing new generation vaccines. Nature Reviews Immunology Vol. 1 No 3: Pages 209-219.
Boerlin, P., S. A. McEwen, F. Boerlin-Petzold, J. B. Wilson, R. P. Johnson and C. L. Gyles (1999). Associations between virulence factors of Shiga toxin-producingEscherichia coli and disease in humans. Journal of Clinical Microbiology Vol. 37 No 3: Pages 497-503.
Brown, J., Willcox, S.J., Franklin, N., Hazelton, B. and O’Sullivan, M.V.N., (2016). Shigellosis: high rates of antibiotic resistance necessitate new treatment recommendations. Medical Journal of Australia Vol. 204 No 7.
Chen, X., J. L. Zaro and W.-C. Shen (2013). Fusion protein linkers: property, design and functionality. Advanced drug delivery reviews Vol. 65 No 10: Pages 1357-1369.
Davies, M. N. and D. R. Flower (2007). Harnessing bioinformatics to discover new vaccines. Drug discovery today Vol. 12 No 9: Pages 389-395.
Doytchinova, I. A. and D. R. Flower (2007). VaxiJen: a server for prediction of protective antigens, tumour antigens and subunit vaccines. BMC bioinformaticsVol. 8 No 1: Page 4.
EL‐Manzalawy, Y., D. Dobbs and V. Honavar (2008). Predicting linear B‐cell epitopes using string kernels. Journal of molecular recognition Vol. 21 No 4: Pages 243-255.
Garnier, J., J.-F. Gibrat and B. Robson (1996). GOR method for predicting protein secondary structure from amino acid sequence. Methods in enzymology Vol. 266: Pages 540-553.
Garnier, J., D. Osguthorpe and B. Robson (1978). Analysis of the accuracy and implications of simple methods for predicting the secondary structure of globular proteins. Journal of molecular biology Vol. 120 No1: Pages 97-120.
Guarner, F., A. G. Khan, J. Garisch, R. Eliakim, A. Gangl, A. Thomson, J. Krabshuis, T. Lemair, P. Kaufmann and J. A. de Paula (2012). World gastroenterology organisation global guidelines: probiotics and prebiotics october 2011. Journal of clinical gastroenterology Vol. 46 No 6: Pages 468-481.
Hale, T. L. (1991). Genetic basis of virulence in Shigella species. Microbiological reviews Vol. 55 No 2: Pages 206-224.
Haste Andersen, P., M. Nielsen and O. Lund (2006). Prediction of residues in discontinuous B‐cell epitopes using protein 3D structures. Protein Science Vol. 15 No 11: Pages 2558-2567.
Hesaraki, M., S. Saadati, H. Honari, G. Olad, R. Ranjbar, M. Heiat and M. Tat (2011). Induction of immune response against ipaD N-terminal region of shigella dysenteriae in guinea pigs. Genetics in the 3rd millennium Vol. 9 No 1: Pages 2261-2266.
Hooft, R., G. Vriend, C. Sander and E. E. Abola (1996). Errors in protein structures. Nature Vol. 381 No 6580: Pages 272-272.
Källberg, M., H. Wang, S. Wang, J. Peng, Z. Wang, H. Lu and J. Xu (2012). Template-based protein structure modeling using the RaptorX web server. Nature protocols Pagesol. 7 No 8: Pages 1511-1522.
La Ragione, R. M., S. Patel, B. Maddison, M. J. Woodward, A. Best, G. C. Whitelam and K. C. Gough (2006). “Recombinant anti-EspA antibodies block Escherichia coli O157: H7-induced attaching and effacing lesions in vitro.” Microbes and infection Vol. 8 No 2: Pages 426-433.
Li, Y., E. Frey, A. M. Mackenzie and B. B. Finlay (2000). Human Response to Escherichia coliO157: H7 Infection: Antibodies to Secreted Virulence Factors. Infection and immunity Vol. 68 No 9: Pages 5090-5095.
Liu, L., H. Johnson, S. Cousens, J. Perin, S. Scott, J. Lawn, I. Rudan, H. Campbell, R. Cibulskis and M. Li (2012). Child Health Epidemiology Reference Group of WHO and UNICEF Global, regional, and national causes of child mortality: an updated systematic analysis for 2010 with time trends since 2000. Lancet Vol. 379 No 9832: Pages 2151-2161.
Lovell, S.C., Davis, I.W., Arendall, W.B., de Bakker, P.I., Word, J.M., Prisant, M.G., Richardson, J.S. and Richardson, D.C., (2003). Structure validation by Cá geometry: ö, ø and Câ deviation. ProteinsVol. 50: Pages 437-450.
Lucas, M. (2001). A reconsideration of the evidence for Escherichia coli STa (heat stable) enterotoxin‐driven fluid secretion: a new view of STa action and a new paradigm for fluid absorption. Journal of applied microbiology Vol. 90 No 1: Pages 7-26.
Lund, O. (2005). Immunological bioinformatics, MIT press.
Ménard, R., P. J. Sansonetti and C. Parsot (1993). Nonpolar mutagenesis of the ipa genes defines IpaB, IpaC, and IpaD as effectors of Shigella flexneri entry into epithelial cells. Journal of bacteriology Vol. 175 No 18: Pages 5899-5906.
Oriá, R.B., Murray-Kolb, L.E., Scharf, R.J., Pendergast, L.L., Lang, D.R., Kolling, G.L. and Guerrant, R.L. (2016). Early-life enteric infections: relation between chronic systemic inflammation and poor cognition in children.Nutrition reviews, p.nuw008.
Paton, J. C. and A. W. Paton (1998). Pathogenesis and diagnosis of Shiga toxin-producing Escherichia coli infections. Clinical microbiology reviews Vol. 11 No 3: Pages 450-479.
Pinheiro, C. S., V. P. Martins, N. R. Assis, B. C. Figueiredo, S. B. Morais, V. Azevedo and S. C. Oliveira (2011). Computational vaccinology: an important strategy to discover new potential S. mansoni vaccine candidates. BioMed Research International Vol. 2011.
Pollastri, G. and A. Mclysaght (2005). Porter: a new, accurate server for protein secondary structure prediction. Bioinformatics Vol. 21 No 8: Pages 1719-1720.
Puigbo, P., E. Guzman, A. Romeu and S. Garcia-Vallve (2007). OPTIMIZER: a web server for optimizing the codon usage of DNA sequences. Nucleic acids research Vol. 35 (suppl 2): Pages W126-W131.
Qadri, F., S. K. Das, A. Faruque, G. J. Fuchs, M. J. Albert, R. B. Sack and A.-M. Svennerholm (2000). Prevalence of toxin types and colonization factors in enterotoxigenic Escherichia coli isolated during a 2-year period from diarrheal patients in Bangladesh. Journal of clinical microbiology Vol. 38 No 1: Pages 27-31.
Rammensee, H.-G., J. Bachmann, N. P. N. Emmerich, O. A. Bachor and S. Stevanovic (1999). SYFPEITHI: database for MHC ligands and peptide motifs. ImmunogeneticsVol. 50 No 3-4: Pages 213-219.
Rodríguez, L., E. Cervantes and R. Ortiz (2011). Malnutrition and gastrointestinal and respiratory infections in children: a public health problem. International journal of environmental research and public health Vol. 8 No 4: Pages 1174-1205.
Roy, A., A. Kucukural and Y. Zhang (2010). I-TASSER: a unified platform for automated protein structure and function prediction. Nature protocols Vol. 5 No 4: Pages 725-738.
Saha, S. and G. Raghava (2004). BcePred: prediction of continuous B-cell epitopes in antigenic sequences using physico-chemical properties. Artificial immune systems, Springer: Pages 197-204.
Sharma, D., Patel, S., Padh, H. and Desai, P., (2016). Immunoinformatic Identification of Potential Epitopes Against Shigellosis. International Journal of Peptide Research and Therapeutics, Pages 1-15.
Sixma, T. K., K. H. Kalk, B. A. van Zanten, Z. Dauter, J. Kingma, B. Witholt and W. G. Hol (1993). Refined structure of Escherichia coli heat-labile enterotoxin, a close relative of cholera toxin. Journal of molecular biology Vol. 230 No 3: Pages 890-918.
Sixma, T. K., S. E. Pronk, K. H. Kalk, E. S. Wartna, B. A. van Zanten, B. Witholt and W. G. Hoi (1991). Crystal structure of a cholera toxin-related heat-labile enterotoxin from E. coli.
Sturniolo, T., E. Bono, J. Ding, L. Raddrizzani, O. Tuereci, U. Sahin, M. Braxenthaler, F. Gallazzi, M. P.
Protti and F. Sinigaglia (1999). Generation of tissue-specific and promiscuous HLA ligand databases using DNA microarrays and virtual HLA class II matrices. Nature biotechnology Vol. 17 No 6: Pages 555-561.
Svennerholm, A.-M. and J. Tobias (2008). Vaccines against enterotoxigenic Escherichia coli.
Taxt, A., R. Aasland, H. Sommerfelt, J. Nataro and P. Puntervoll (2010). Heat-stable enterotoxin of enterotoxigenic Escherichia coli as a vaccine target. Infection and immunity Vol. 78 No 5: Pages 1824-1831.
Taxt, A.M., Diaz, Y., Aasland, R., Clements, J.D., Nataro, J.P., Sommerfelt, H. and Puntervoll, P., (2016). Towards rational design of a toxoid vaccine against the heat-stable toxin of Escherichia coli. Infection and immunity Vol. 84 No 4: Pages 1239-1249.
Wiederstein, M. and M. J. Sippl (2007). ProSA-web: interactive web service for the recognition of errors in three-dimensional structures of proteins. Nucleic acids research Vol. 35 (suppl 2): Pages W407-W410.
Yang, J., R. Yan, A. Roy, D. Xu, J. Poisson and Y. Zhang (2015). The I-TASSER Suite: protein structure and function prediction. Nature methods Vol. 12 No 1: Pages 7-8.
Zeinalzadeh, N., A. H. Salmanian, G. Ahangari, M. Sadeghi, J. Amani and M. Jafari (2013). Designing and production of a chimeric multi-epitope construct of CfaB, ST toxoid, CssA, CssB and LTB against enterotoxigenic E. coli. Current Opinion in Biotechnology No 24: S106.
Zhang, W., C. Zhang, D. H. Francis, Y. Fang, D. Knudsen, J. P. Nataro and D. C. Robertson (2010). Genetic fusions of heat-labile (LT) and heat-stable (ST) toxoids of porcine enterotoxigenic Escherichia coli elicit neutralizing anti-LT and anti-STa antibodies. Infection and immunity Vol. 78 No 1: Pages 316-325.
Zuker, M. (2003). Mfold web server for nucleic acid folding and hybridization prediction. Nucleic acids research Vol. 31 No 13: Pages 3406-3415.