*Department of Botany, MES Abasaheb Garware College, Pune 4, MS, India
1Department of Botany, Modern College of Arts, Science and Commerce, Pune 5, MS, India
Corresponding author email: gnivedita_ghayal@rediffmail.com
Article Publishing History
Received: 18/04/2020
Accepted After Revision: 27/05/2020
Cassia uniflora Mill. non Spreng.and Synedrella nodiflora (L.) Gaertn. are invasive alien weeds on Deccan plateau. Both the weeds are spreading on agricultural and fallow lands at an alarming rate replacing many native and exotic species. Along with the detail studies of their morpho-physiological, metabolomic and chemical attributes, an attempt was made to explore their ecological survival and dominance, which is responsible for development of huge monothickets. Their invasion success probably is due to containment of similar ecological behaviour and evolutionary relatedness to potential plant invaders. In the present study we have extensively employed different tools of bioinformatics such as BLAST, FASTA, servers like SWISSMODEL, CLUSTAL OMEGA and software like MEGA-X, and carried out phylo-genomics and evolutionary cladistic analyses for protein structures of important enzymes such as Rubisco and Maturase –K. The selected weeds were compared using molecular data from gene sequences with each other and other co-dominant native and exotic species. The results revealed that all the species under focus shared larger part (80%) of MSAs (Multiple Sequence Alignments). The data indicated that Cassia uniflora and Synedrella nodiflora exhibited parallel resistance to the environmental stresses, similar evolutionary patterns and highlighted their dominance amongst different species of Cassia and respective genera of Asteraceae. Based on further phylogenetic studies it can be proposed that C. auriculata and Lactuca indica would be the future successful invaders on Deccan plateau. The present investigation based on the results of MSA, deep view analyses and phylogenomics of weeds may predict the changes in weed flora of Deccan pleateau due to environmental changes.
Invasive weeds, Maturase-K, MEGA-X, Phylogeny, Rubisco
Ghayal N, Deodhar A. Cladistic Analysis and Comparative Account of Different Invasive Weeds and their Dominance Using Different Bioinformatics Tools. Biosc.Biotech.Res.Comm. 2020;13(2).
Ghayal N, Deodhar A. Cladistic Analysis and Comparative Account of Different Invasive Weeds and their Dominance Using Different Bioinformatics Tools. Biosc.Biotech.Res.Comm. 2020;13(2). Available from: https://bit.ly/2RU6kel
Copyright © Ghayal and Deodhar This is an open access article distributed under the terms of the Creative Commons Attribution License (CC-BY) https://creativecommns.org/licenses/by/4.0/, which permits unrestricted use distribution and reproduction in any medium, provide the original author and source are credited.
INTRODUCTION
The alien species highly out-compete the native species or escape from adverse environmental conditions and dominate the community (MacDougall and Turkington 2005).Their diversity is controlled by population, ecosystem dynamics, disturbances, nutrient supply and climatic factors. The biotic restrictions also force them to skip from their previous habitat and start surviving in new habitats, helping in the process of invasion (Mack et al. 2000). Many a times these phyto-invasives become very aggressive due to production of some defensive chemicals (Carpenter and Cappuccino 2005). These invasions pose many ecological, economic and social problems. Because of this the studies on plant invasions and its mechanism and consequences of them on global biodiversity and ecosystem functioning are of urgent need. This is because slowly and gradually these invasives become aggressive and encroach cultivable lands and pose a great problem (Chauhan et al. 2017).
Cassia uniflora Mill. non Spreng. (family- Caesalpinaceae) is annual, erect herb with yellow flowers and clustered pods. It originated in tropical South America and now distributed worldwide. This invasive weed grows luxuriantly at many places (Almeida, 2003). Synedrella nodiflora (L.) Gaertn. (family- Asteraceae) originated in tropical America, is an annual, erect, dichotomously branched herb distributed all over India (Almeida, 2003). Their dominance is attributed to wide adaptability to diverse habitat, different morpho-physiological characters and defensive allelo-chemicals (Ghayal et al.2007a,b; 2009; 2013).
Limited research has been done on the allelopathic effect or phyto-toxicity of Cassia uniflora to other plants. There is a general temper of agreement now-a-days that invasive plants displace the local biodiversity through their harmful effects including allelopathy (Cronk and Fuller, 1995). Allelopathic effects may due to the presence of allelochemicals in Cassia and Synedrella, like different types of phenolic compounds, alkaloids, triterpenoides, essential oils and flavonoids, biocides, juvenile hormones, growth hormones. They may be interacting with various physiological processes (Chatterjee et. al. 2012). From current literature review, it revealed that little research has been done on distribution, evolutionary studies and impact of Cassia uniflora and Synedrella nodiflora on co-occuring species by using various tools of bioinformatics. The work done till now on the metabolic compounds and various allelochemicals has been restricted only to the wet lab methods and very little is known about the gene level expression of all such compounds. Some advanced researches show the gene expression analysis on different weeds that indicate several compounds responsible for the invasion of weeds into new environments ( Chen, 2013).
For bioinformatics study, several soft-wares and tools were used to analyse the data present on both the weeds. Due to lack of research on the weed plants, it was difficult to retrieve the molecular data. Hence the common enzymes in Cassia uniflora Mill. non Spreng, and Synedrella nodiflora (L) Gaertn, were selected for further analysis. The enzymes or proteins that were studied in both the invasive weeds are – Maturase K [EC 2.7.10.2] and Ribulose-1,5-bisphosphate carboxylase / oxygenase(Rubisco/ rbcL)[EC 4.1.1.39]. Maturase K is a plant plastidial gene. The protein it encodes is an intron Maturase, a protein that splices introns. Mat-K is proposed as the only chloroplast-encoded group II intron Maturase, thus implicating Mat-K in chloroplast posttranscriptional processing. For a protein-coding gene, mat-K has an unusual evolutionary significance, including relatively high substitution rates at both the nucleotide and amino acids levels, (Barthet et. al.2015).
The other enzyme that has been studied in both plants was ribulose-1,5-bisphosphate carboxylase/oxygenase (Rubisco) is the major enzyme assimilating CO2 into the biosphere. At the same time Rubisco is an extremely inefficient catalyst and its carboxylase activity is compromised by an opposing oxygenase activity involving atmospheric O2. These enzymes were considered for checking the probable similarity in the protein sequences of these two weeds. The nucleotide research on these enzymes is done intensively since both the enzymes play crucial role in plant metabolism. The focus of the study was to acquire the common factor based on genomic data in both the weeds that is responsible for their dominance and to understand and interpret their ecological and evolutionary significance.
The present work was carried out for correlating different genes or proteins in Cassia uniflora Mill. non Spreng and Synedrella nodiflora (L) Gaertn. responsible for their invasiveness by using different bioinformatics tools. Scanty information is available on the molecular level work, gene identification and sequencing of the invasive weeds as compared to the crop plants. Therefore present attempt was made to fulfil this gap in above mentioned weeds. It has also helped to get the idea about the ecological corridor developed by current dominant invasive weeds. Not only that but it has also helped to predict the future changes in weed flora and their ecological status.
MATERIAL AND METHODS
- A) By performing Multiple Sequence Alignment (MSA):The Multiple Sequence Alignment of the 2 or more sequences was done to check whether the sequences align exactly similar to each other. Here, sequence homology is applied to assess if the sequences are sharing evolutionary origins. B) By using ‘Deep-view software’: This software was used for direct analysis of the similar or distant proteins. The proteins were modelled from the server ‘SWISSMODEL’ in PDB format to get the 3D structure. And then these proteins were analysed in the software. C) Preparation of phylogenetic tree: It was carried out to check the homology between the selected protein sequences. Cladistics analysis was performed by using software MEGA-X with the help of maximum likelihood method. Website used – ncbi.nlm.nih.gov
RESULTS AND DISCUSSION
For tracking down the upshots on invasive species Cassia uniflora/ Senna uniflora and Synedrella nodiflora the efforts were carried out in the following manner –A) By performing Multiple Sequence Alignment (MSA) – For Maturase-K and Rubisco for Cassia uniflora/ Senna uniflora and Synedrella nodiflora. B) By using ‘Deep-view software’- For protein structure of Maturase-K and Rubisco of Cassia uniflora/ Senna uniflora and Synedrella nodiflora. C) Preparation of phylogenetic tree / Cladistic analyses
C) Preparation of phylogenetic tree / Cladistic analyses –
Table 1. Phylogenetic trees of invasive species Cassia and Synedrella
| No. | Type of
Phylogenetic tree |
Enzyme | MSA % |
| 1. | Tree and herb species of Cassia/ Senna | Maturase-K | 90% |
| 2. | Tree and herb species of Cassia/ Senna | Rubisco | Negligible – the authenticity of this clad was very low and hence was not considered for comparison |
| 3. | Weed species of Cassia/ Senna | Maturase-K | 95% |
| 4. | Weed species of Cassia/ Senna | Rubisco | 90% |
| 5. | Synedrella nodiflora and other weed species | Maturase-K | 60% |
| 6. | Synedrella nodiflora and other weed species | Rubisco | 90% |
| 7. | Cassia/ Senna, Synedrella and other related genera and species | Maturase-K | 80% |
| 8. | Cassia/ Senna, Synedrella and other related genera and species | Rubisco | 85-90% |
- A) Results of Multiple Sequence Alignment (MSA):Multiple sequence alignment showed that the sequence of enzyme ‘Maturase-K’ is exactly similar in plants Cassia unifloraand Synedrellanodiflora. This result revealed the sequences with almost homologous regions showing ‘*’symbol as exactly matching sequences (Fig.1, Table 2). Similar MSA was carried out on Rubisco for the same two plants Cassia uniflora and Synedrella nodiflora and equivalent similarity was observed. The figure and table for this are not included to avoid repetition of the sets.
A) By Multiple Sequence Alignment (MSA)
MSA of Cassia uniflora and Synedrella nodiflora for Maturase-K enzyme-
Figure 1: Showing Multiple Sequence Alignment for Maturase-K enzyme
Table 2. Showing details of two weeds used for MSA of Maturase-K enzyme
| Name of the Plant | Accession Number | Number of Base Pairs
(Amino acids) |
Included or
Excluded |
MSA percentage of Sequence Similarity
All these species show about 70 % sequence similarity |
| Cassia uniflora | ARR68700.1 | 280 | Included | |
| Synedrella nodiflora | AAR02837.1 | 508 | Included |
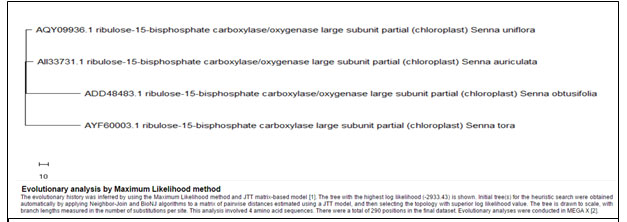
When similar MSA (Multiple sequence alignment/s) was carried out for Rubisco enzyme, Cassia auriculata had to beexcepted because it has different base pairs than all other plants which was affecting the comparison among the selected plants. The comparison of remaining plants showed almost similar sequences in all the species related to the weed species under consideration i.e. Cassia uniflora and Synedrella (Fig. 2, Table 3). Similar comparison is done for Maturase-K enzyme and it shows similar results except the inclusion of Cassia auriculata. Hence it can be predicted that these invasive species could have evolved in similar way for the enzymes like ‘Maturase-K’ and Rubisco.
MSA for all related species of both Cassia uniflora and Synedrella nodiflora for Rubisco enzyme –
Figure 2: Showing Multiple Sequence Alignment for Rubisco
Table 3: Showing details of nine plants used for MSA of Rubisco enzyme
| Name of the Plant | Accession Number | Number of Base Pairs (Amino acids) | Included or Excluded | MSA percentage of Sequence Similarity |
| Cassia uniflora | AQY09936.1 | 249 | Included | All these species together show about 80 % sequence similarity |
| Cassia obtusifolia | ADD48483.1 | 234 | Included | |
| Cassia tora | AYF60003.1 | 238 | Included | |
| Cassia auriculata | AII33731.1 | 290 | Excluded | |
| Cassia occidentalis | AZC11295.1 | 416 | Included | |
| Cassia sophera | AFU54416.1 | 202 | Included | |
| Synedrella nodiflora | ARR11741.1 | 468 | Included | |
| Tridax procumbens | AFP23718.1 | 461 | Included | |
| Pulicaria dysenterica | AKG25301.1 | 439 | Included |
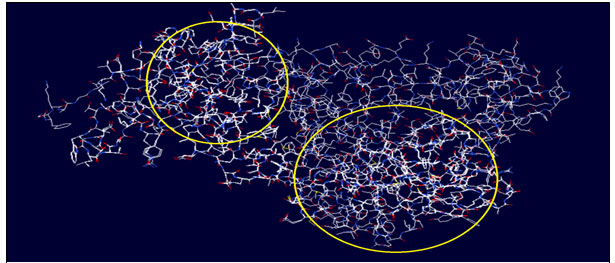
Results of using ‘Deep-View’ software: When the models of protein structures for both the enzymes ‘Maturase-K’ and ‘Rubisco’ were run in Deep view software, models exhibited similar sequences of both the enzymes that implied the similar structures and hence all the models tracked in the software were able to get merged. Results of deep view analysis (Fig. 3 & 4) showed that the enzymes studied had identical structures and the highlighted portion in both the images showed shared structures of the same protein but in two different plants Cassia / Senna unifloraand Synedrella. This probable further confirms simultaneous evolution of these invasive species and their imperative enzymes.
Figure 3: Circled portion indicates common sequence coding similar structure of enzyme MATURSE-K in Cassia uniflora and Synedrella nodiflora
Figure 4: Circled portion indicates common sequence coding similar structure of enzyme RUBISCO in Cassia uniflora and Synedrella nodiflora
Preparation of phylogenetic trees / Cladistic analyses: The construction of phylogenetic trees was carried out with the interest of searching the evolutionary associations of Cassia/ Senna uniflorawith the other native, weedy and less dominant species of Cassia. In case of Synedrella searching was attempted with the same approach but due to very much insubstantial outcomes, the cladograms were developed based on few weedy, local and fairly prevailing genera and species from the same family Asteraceae, as this family is well known to contain globally distributed, highly dominant invasive weed genera other than Synedrella.
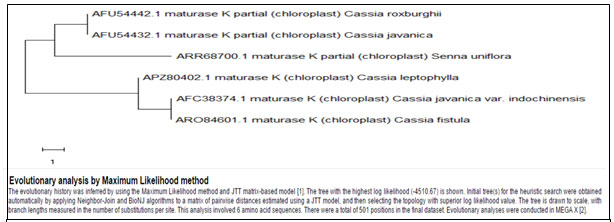
The Cladistic analysis (Fig. 5, Table 4) performed on different herb and tree species of Cassia revealed that Cassia uniflora had a distant phylogeny and did not share any recent ancestry with any of the other Cassia species for Maturase-K enzyme.As against that when the phylogenetic comparison of different weed species of Cassia / Senna was carried out for these two enzymes, it was observed that Cassia unifloraand Cassia auriculatamight have progressed in the most parallel way and from very recent common ancestor in due course of evolution (Figures 6, 7; Tables 5,6). It can also be confirmed by the habitat of both the species of Cassia. Both Cassia uniflora and Cassia auriculata grow intensively in semi-arid conditions. The similar phylogeny here, in both the enzymes, Maturase-K and Rubisco, suggests that these two plants must have acquired similar properties that help them survive under the unfavourable conditions and this could be the key to their dominance over other native plants. The results showed that the evolution of Cassia uniflora and Cassia auriculata is relatively similar to each other over other species of Cassia. Cassia uniflora is the most dominant species of the genus Cassia presently and Cassia auriculata might evolve further as dominant species in subsequent situations of environment, as Cassia uniflora is today. The other species of Cassia such as Cassia occidentalis and Cassia tora have shown distant phylogeny. Both Cassia uniflora and Cassia auriculata are distantly evolved from other species of Cassia and hence show significantly different habitat and possibly also the chemical properties than other members of genus Cassia.
- Preparation of phylogenetic tree / Cladistic analyses
Cladistics analysis between different species ofCassia for Maturase-K enzyme-
Figure 5: Comparison between different species of Cassia for maturase-K enzyme
Table 4. Showing details of different species of Cassia for Maturase-K enzyme
| Name of the Plant | Accession Number | Number of Base Pairs (Amino acids) | Included or Excluded | MSA percentage of Sequence Similarity
All these species show about 90 % sequence similarity |
| Cassia uniflora | ARR68700.1 | 280 | Included | |
| Cassia roxburghii | AFU54442.1 | 278 | Included | |
| Cassia javanica | AFU54432.1 | 278 | Included | |
| Cassia leptophylla | APZ80402.1 | 499 | Included | |
| Cassia javanica var. indochinensis | AFC38374.1 | 501 | Included | |
| Cassia fistula | ARO84601.1 | 501 | Included |
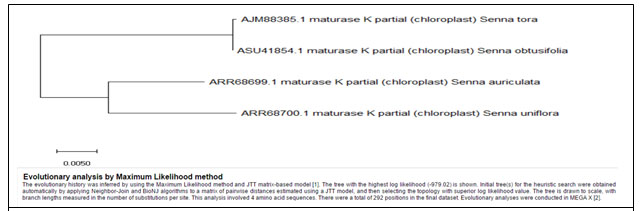
Cladistics analysis between weed species of Cassia for Maturase-K enzyme–
Figure 6: Showing Phylogenetic tree of four weed species for Maturase-K enzyme
Table 5. Showing details of four weed species of Cassia used for MSA of Maturase-K enzyme
| Name of the Plant | Accession Number | Number of Base Pairs (Amino acids) | Included or Excluded | MSA percentage of Sequence Similarity
All these species show about 95 % sequence similarity |
| Cassia uniflora | ARR68700.1 | 280 | Included | |
| Cassia obtusifolia | ASU41854.1 | 276 | Included | |
| Cassia tora | AJM88385.1 | 248 | Included | |
Cassia auriculata |
ARR68699.1 | 261 | Included |
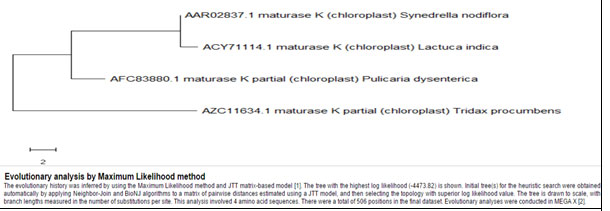
The comparison of Synedrella in cladogram with the other genera of asteraceae for Maturase-K showed that, number of base pairs varies greatly among genera and hence the MSA percentage has fallen (60%) but the matching base pairs show completely identical sequences for all the plants. Synedrella nodiflora indicated distant phylogeny from Tridax procumbens and Pulicaria dysenterica but is relatively nearer to Lactuca indica (Fig. 8, Table 7). These three genera of Asteraceae show their frequent but less dominant occurrence than Synedrella.For the same members of asteraceae, the building up of phylogenetic tree for Rubisco was performed excluding Lactuca indica as its protein sequence was unavailable. Here, it was observed that Tridax procumbens and Pulicaria dysenterica had the closest phylogeny showing Synedrella nodiflora distantly placed (Fig. 9, Table 8).
Figure 7: Showing phylogenetic tree of weedspecies for Rubisco enzyme
Table 6. Showing details of weedspecies for Rubisco enzyme
| Name of the Plant | Accession Number | Number of Base Pairs (Amino acids) | Included or Excluded | MSA percentage of Sequence Similarity
All these species show about 90 % sequence similarity |
| Cassia uniflora | AQY09936.1 | 249 | Included | |
| Cassia obtusifolia | ADD48483.1 | 234 | Included | |
| Cassia tora | AYF60003.1 | 238 | Included | |
Cassia auriculata |
AII33731.1 | 290 | Excluded |
Cladistics analysis for Synedrella nodiflora withherb species of Asteraceae family members for Maturase-K enzyme
Figure 8: Showing Phylogenetic tree of herb species of members of Asteraceae for Maturase-K enzyme
Table 7. Showing details of asteraceae members herb species for Maturase-K enzyme
| Name of the Plant | Accession Number | Number of Base Pairs
(Amino acids) |
Included or
Excluded |
MSA percentage of Sequence Similarity
All these species show about 60 % sequence similarity |
| Synedrella nodiflora | AAR02837.1 | 506 | Included | |
| Tridax procumbens | AZC11634.1 | 254 | Included | |
| Lactuca indica | ACY71114.1 | 506 | Included | |
Pulicaria dysenterica |
AFC83880.1 | 324 | Included |
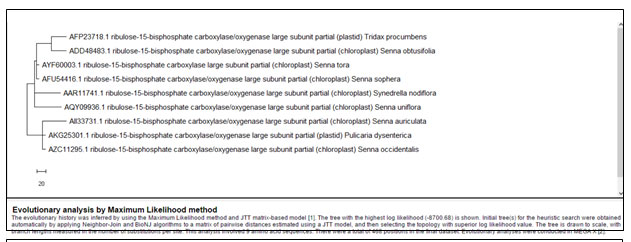
The comparison of both Cassia uniflora and Synedrella nodiflora for Maturase-K showed very distant origin. Cassia uniflora showed quite distant phylogeny from other Cassia species (Fig. 10, Table 9) whereas; Synedrella nodiflora shares similar phylogeny with Lactuca indica. In contrast the comparison for Rubisco showed common ancestry for Cassia uniflora and Synedrella nodiflora as they are placed very close to each other (Fig. 11, Table 10). This shows that both the plants might have developed different evolutionary patterns than all the other plants under consideration which resulted in distant phylogeny of both. In this clad, Lactuca indica was not considered for the comparison as its protein sequence for Rubisco was unavailable (Fig. 11, Table 10). The importance of genomic tactics for understanding the weedy and invasive behaviours of plants, their evolution and resistant response to environmental fluctuations is better realised now as a part of weed biology ( Stewart et al. 2009).
Cladistics analysis for Synedrellanodiflorawith herb species of Asteraceae family members for Rubisco enzyme –
Figure 9: Showing Phylogenetic tree of herb species of members of Asteraceae for Rubisco enzyme
Table 8: Showing details of herb species of members of Asteraceae for Rubisco enzyme
Table 8. Showing details of herb species of members of Asteraceae for Rubisco enzyme
| Name of the Plant | Accession Number | Number of Base Pairs
(Amino acids) |
Included or
Excluded |
MSA percentage of Sequence Similarity
All these species show about 90 % sequence similarity |
| Synedrella nodiflora | AAR11741.1 | 468 | Included | |
| Tridax procumbens | AFP23718.1 | 461 | Included | |
| Lactuca indica | – | – | Excluded | |
Pulicaria dysenterica |
AKG25301.1 | 439 | Included |
Cladistics analysis for weed related species of both Cassia uniflora and Synedrella nodiflora for Maturase-K enzyme –
Figure 10: Showing Phylogenetic tree of all 9 species for Maturase-K enzyme
Table 9. Showing details of weedspecies for Maturase-K enzyme
| Name of the Plant | Accession Number | Number of Base Pairs
(Amino acids) |
Included or
Excluded |
MSA percentage of Sequence Similarity
All these species together show about 80 % sequence similarity |
| Cassia uniflora | ARR68700.1 | 280 | Included | |
| Cassia obtusifolia | ASU41854.1 | 276 | Included | |
| Cassia tora | AJM88385.1 | 248 | Included | |
| Cassia auriculata | ARR68699.1 | 261 | Included | |
| Cassia occidentalis | AZC11603.1 | 269 | Included | |
| Cassia sophera | AFU54436.1 | 278 | Included | |
| Synedrella nodiflora | AAR02837.1 | 506 | Included | |
| Tridax procumbens | AZC11634.1 | 254 | Included | |
| Lactuca indica | ACY71114.1 | 506 | Included | |
Pulicaria dysenterica |
AFC83880.1 | 324 | Included |
It further opens a new research route for perception of reckless growth and evolution of phyto-invasives and their functioning under harsh stress conditions. It also shares knowledge about weed management, herbicide resistance mechanism of allelopathy and evolution of invasiveness of such plant species (Thomas and Klaper, 2004). Such phylogenetic studies using various methods help to better understand causes of invasion success, ecosystem disturbance and alterations in biodiversity (Forest et al., 2007; Proches et al., 2008; Winter et al., 2009; Dawson et al., 2009).
Cladistics analysis for weed related species of both Cassia uniflora and Synedrella nodiflora for Rubisco enzyme –
Figure 11: Showing Phylogenetic tree of all 9 species for Rubisco enzyme
Table 10. Showing details of weed species for Rubisco enzyme
| Name of the Plant | Accession Number | Number of Base Pairs
(Amino acids) |
Included or
Excluded |
MSA percentage of Sequence Similarity
All these species together show about 90 % sequence similarity |
| Cassia uniflora | AQY09936.1 | 249 | Included | |
| Cassia obtusifolia | ADD48483.1 | 234 | Included | |
| Cassia tora | AYF60003.1 | 238 | Included | |
| Cassia auriculata | AII33731.1 | 290 | Included | |
| Cassia occidentalis | AZC11295.1 | 416 | Included | |
| Cassia sophera | AFU54416.1 | 202 | Included | |
| Synedrella nodiflora | AAR11741.1 | 468 | Included | |
| Tridax procumbens | AFP23718.1 | 461 | Included | |
| Lactuca indica | – | – | Excluded | |
Pulicaria dysenterica |
AKG25301.1 | 439 | Included |
Some studies have also claimed that phylogenetic and functional attributes of alien species readdress different aspects of ecosystem functioning and variations produced at the level of organisms (Chen, 2013; Ricotta et al. 2009, 2010; Cadotte et al. 2009). According to some researchers unusual characters and ancestral relations with natives probably are promoting the aliens to become invasive very swiftly in non-native ranges of global vegetation (Clements and Ditommaso, 2011).Bezeng et al. (2013) have claimed that phylo-genomic studies of invasive species with reference to their native co-survivors are the most important drivers of ecosystem change, which can alter the vegetational set up of a particular area. Similar studies on Maturase-K and Rubisco of island flora have been carried out by them to understand the causes of invasion on Robben island, South Africa. The results recorded in the figures 1 and 2 and Tables 2 and 3 showed that multiple sequence alignments are shared for Maturase-K and Rubisco of Cassia uniflora and Synedrella. Deep view analysis (Figures 3 and 4) also revealed major portions identical for both the enzyme proteins for C. uniflora and Synedrella. This further indicates that these two enzymes might be the drivers in the invasion success of C. uniflora and Synedrella(Bezeng et al., 2013), since these enzymes have prime importance in the plant metabolism.
When evaluation of different species of Cassia for Maturase-K was carried out (Fig. 5, Table 4), it exhibited minor likelihood of C. uniflora from others. Further, when we examined the cladistics patterns of different species of Cassia for both the enzymes (Figures 6, 7 and Tables 5, 6), it was observed that C. auriculata is lineally related to C. uniflora, indicating similar or parallel functional traits which are essential for invasion. Martyniuk et al. (2009) have phyletically compared members of Amaranthaceae in the same way for pollen structures based on these two enzymes.The evaluation of different asteraceae members along with the invasive alien species Synedrellanodiflorasuggested its racial link with Lactucaindica for Maturase-K (Fig.8, Table 7). When Synedrellanodiflorawas (Fig. 9, Table 8) compared with same herb species from asteraceae for Rubisco revealed distant phylogeny. The consideration for phyletic relatedness when was performed for Maturase-K (Fig. 10, Table 9), showed totally separate placement of C. uniflora but indicating probable common ancestry. This clad suggested co-evolution of Synedrella nodiflora and Lactuca indica. The same set of plants was used to prepare clad for Rubisco revealed (Fig. 11, Table 10) the diversification from the common inherited line. The studies on evolutionary population genomics in the Asteraceae family have been carried out by Stevens (2007), Barker et al. (2008), Broz et al. (2007) and Mandel et al. (2017).
Thus the present study has facilitated to establish the correlations of the invasive species Cassia uniflora and Synedrella nodiflora with the other innate and aggressive species either of the same genus or family. The phylogenomic studies using bio-computing tools enabled to understand the protein nature of Maturase-K and Rubisco of these two weeds mainly and also of other species. There could be generated various clades giving insights into the evolutionary connections of these aliens developing monothickets with each other along with the other plants. Further it will help to know the changing and dominating weed flora in the same area.
CONCLUSION
Overall this research work points out to the protein based phylogenetic similarities and distinctiveness of alien taxa with respect to the other genera as significant details deciding their invasion success (Ordonez, 2014). This enhances to the idea of phylogenetic and metabolic patterns of successfully invaded species. Further these studies have focussed light on the future invasive followers of them on Deccan plateau through different probability patterns.
ACKNOWLEDGEMENTS
The authors are thankful to Principal and Head, Department of Botany, MES Abasaheb Garware College, Pune and Modern College of Arts, Science and Commerce, Shivajinagar, Pune, for their constant support in carrying out this research.
REFERENCES
Almeida M.R., Dutta. S, and Almeida S. M.(2003) Glimpses of Phytogeography of Maharashtra. Journal of Bombay Natural History SocietyVol.100 ; Pages 559 – 5885
Barker M. S., Kane N. C, Kozik A, Michelmore R. W, Matvienko M. (2008) Multiple paleopolyploidizations during the evolution of the Asteraceae reveal parallel patterns of duplicate gene retention after millions of years. Molecular Biology andEvolution 25: Pages 2445–2455
Barthet M. M., Keenan. Moukarzel, K N. Smith, Jaimin Patel,and K Whilu. (2015) Alternative translation initiation codons for the plastid maturase Matk unraveling the pseudogene misconception in the Orchidaceae. BMC Evolutionary Biology 15 Page 210 DOI 10.1186/s12862-015-0491-1.
Bezeng S. Bezeng., Vincent Savolainen, KowiyouYessoufou, Alexander S.T. Papadopulos, Olivier Maurinet .(2013) A phylogenetic approach towards understanding the drivers of plant invasiveness on Robben Island South Africa, Botanical Journal of the Linnean Society, Vol.172 : Pages 142–152.
Broz A. K., Broeckling C. D, He J, Dai X, Zhao P. X, (2007) A first step in understanding an invasive weed through its genes: an EST analysis of invasive Centaurea maculosa, BMC Plant Biology Vol.7 Page25. http://www.biomedcentral.com/1471-2229/7/25.
Cadotte M. W., Hamilton M. A, & Murray B. R. (2009) Phylogenetic relatedness and plant invader success across two spatial scales. Diversity and Distributions 15(3): Pages 481–488.
Carpenter D and Cappuccino N. (2005) Herbivory time since introduction and the invasiveness of exotic plants. Journal of Ecology Vol. 93: Pages 315 – 321.
Chatterjee Saheli., Sabyasachi Chatterjee, Aloke Bhattacharya and Sikha Dutta. (2012) Allelopathic Effect of Cassia occidentalis Leaves on Mustard Seeds. Trends in Biotechnology Research 1 No1: Pages 29 -35.
Chauhan S., Amar Matloob, Gulshan Mahajan, Farhena Aslam, S. K. Florentine and Prashant Jha (2017) Emerging Challenges and Opportunities for Education and Research in Weed Science. Front. Plant Sci., Vol.8. Article 1537.
Clements D and Ditommaso A. (2011) Climate change and weed adaptation: can evolution of invasive plants lead to greater range expansion than forecasted? .Weed Research 51No3: Pages 227–240.
Cronk Q. C .B. and Fuller J. L.(1995) Plant Invaders the Threat to Natural Ecosystems, Chapman and Hall, London.
Dawson W, Burslem D.F. and Hulme P. E. (2009) Factors explaining alien plant invasion success in a tropical ecosystem differ at each stage of invasion. Journal of Ecology97: Pages 657 – 665.
Forest F.,Grenyer R, Rouget T.M, Davies J, Cowling R.M.(2007) Preserving the evolutionary potential of floras in biodiversity hotspots. Nature445: Pages757–760.
Ghayal N. A., Dhumal K. N, Deshpande N. R, Shah S. M and Tambe (2007a) Steam Volatile Components from Cassia uniflora and Synedrellanodiflora by Gas Liquid Chromatography – Mass Spectroscopy. Journal of Indian Council of Chemists Vol. 24 No 2 : Pages 60 -62.
Ghayal N. A., Dhumal K. N, Deshpande N. R, Shah S. M and Ruikar A. D.(2007b) Studies on Allelochemicals in Synedrellanodiflora and Impact of its Leaf Leachates on Germination and Seedling growth of Radish (Raphanus sativus) and Mustard (Brassica juncea).Asian Journal of Chemistry 20 No 8 : Pages 6114 – 6120.
Ghayal N. A., Dhumal K. N, Gupta S G, Parange S and Phadke M. (2009) Mophophysiological studies in some invasive weeds from deccan pleateau. Journal of Plant interactions4 No1: Pages 33 – 39. DOI- 10.1080 / 17429140802385964
Ghayal N. A., Dhumal K. N, Deshpande N. R, Ruikar A .D and Usha Phalgune.(2013) Phytotoxic Effects of Leaf Leachates of an Invasive Weed Synedrella nodiflora and characterization of its allelochemicals. International Journal of Pharmacy Science Review and Research 19 No 1: 17: Pages 79-86.
MacDougall A. S and Turkington R. (2005) Are invasive species the drivers or passengers of change in degraded ecosystems? Ecology Vol. 86 : Pages42 – 55.
Mack R. N., Simberloff D, Lonsdale W. M, Evans H and Clout M.(2000) Biotic invasions: Causes, epidemiology, global consequences and control, Ecology Appl.10 : Pages 689–710.
Mandel J. R., M. S. Barker, R. J. Bayer , R. B. Dikow , Tian-Gang Gao , K. E. Jones, S. Keeley, N. Kilian, Hong Ma, C. M. Siniscalchi, A. Susanna, Ramhari Thapa, Linda Watson and V. A. Funk (2017) The Compositae Tree of Life in the age of phylogenomics. Volume 55. No. 4: 405–410.
Martyniuk O. O., Nalian A. G, Van Kley .J. E and Martynova O. V. (2009) Phylogenetic assay of maturase k, ribulose bisphosphate carboxylase (rbcl) sequences, and pollen structure of representatives of the family Amaranthaceae Biotechnologia Acta Vol. 2 No2 : Pages 98 – 103.
Neal Stewart. C. Jr., Tranel P. J, Horvath D. P, Anderson J. V, Rieseberg L. H. (2009) Evolution of Weediness and Invasiveness: Charting the Course for Weed Genomics. Weed Science 57 : Pages 451–462.
Ordonez A. (2014) Functional and Phylogenetic similarity of alien plants to co-occurring Natives. Ecology Vol. 95 No 5: Pages 1191-1202. https://doi.org/10.1890/13-1002.1
S., Wilson J. R. U, Richardson D.M and Rejmánek M. (2008) Searching for phylogenetic pattern in biological invasions. Global Ecology and Biogeography Vol. 17 : Pages 5–10.
Ricotta C., Godefroid S and Rocchini D. (2010) Invasiveness of alien plants in Brussels is related to their phylogenetic similarity to native species, Diversity and Distributions 16 No 4 : Pages 655–662.
Ricotta C., LaSorte F. A, Pysek P, Rapson G. L, Celesti-Grapow L.(2009) Phyloecology of urban alien floras. Journal of Ecology 97 No 6 : Pages 1243–1251.
Stevens P. F. (2007) Angiosperm Phylogeny, Version 8 http://www.mobot.org/MOBOT/research/APweb/Accessed: January 13, 2008.
Thomas M. A. and R Klaper. (2004) Genomics for the ecological toolbox. Trends EcologyE Vol. 19 : Pages 439–445.
Winter M., Schweigera O, Klotza S, Nentwigc W, Andriopoulosd (2009) Plant extinctions and introductions lead to phylogenetic and taxonomic homogenization of the European flora. Proceedings of the National Academy of Sciences of the United States of America Vol. 106 : Pages 21721–21 725.
Youhua Chen. (2013) Distributional Patterns of Alien Plants in China: The Relative Importance of Phylogenetic History and Functional Attributes. ISRN Ecology(527052: 1 -9 Article ID 527052), http://dx.doi.org/10.1155/2013/527052