Department of Computer Science, PSGR Krishnammal College for Women, Peelamedu, Coimbatore, 641004, India
Corresponding author email: gripsyjebs@gmail.com
Article Publishing History
Received: 05/01/2020
Accepted After Revision: 26/02/2020
The identification of specific regions in an organism of a plant, animal or a single-celled life form, or something that has interdependent parts and that is being compared to a living creature. User can choose and select any type of organism’s image such as plant or animal. After successful selection, the proposed tool automatically analyses and finds the name of the organisms and identifies the specific region with some other related information in an effective manner.In comparative and evolutionary genomics, a detailed comparison of common features between organisms is essential to evaluate genetic distance. However, identifying differences in matched and mismatched genes among multiple genomes is difficult using current comparative genomic approaches due to complicated methodologies or the generation of meager information from obtained results. This research reduces the manual activity to analyze the details. Using this software tool, it easily helps us to know all the related organisms information of specific region and also can make report effectively. This makes the system user-friendly consequently reducing the manual work. The system has been developed with advanced features. The objective of this work is to establish an identification of specific region organisms and related information.The system developed with the main intension to progress an effective and user friendly tool for identification of the specific region in organisms. From the experimental results, the proposed system grasps and more and more identification accuracy.
Classification, Organism, Organism Search, Region Detection, Specific Region
Gripsy J. V, Kavitha A. S, Menaka V. Biological Software for Recognition of Specific Regions in Organisms. Biosc.Biotech.Res.Comm. 2020;13(1).
Gripsy J. V, Kavitha A. S, Menaka V. Biological Software for Recognition of Specific Regions in Organisms. Biosc.Biotech.Res.Comm. 2020;13(1). Available from: https://bit.ly/3aqf2aK
Copyright © Gripsy et al., This is an open access article distributed under the terms of the Creative Commons Attribution License (CC-BY) https://creativecommons.org/licenses/by/4.0/, which permits unrestricted use distribution and reproduction in any medium, provide the original author and source are credited.
INTRODUCTION
This communication describes the main event, ie to identify organisms using protein sorting process, which provides an overview of the computational contributions made to this field, and finally gives a few guiding words to potential users. Gene amplification and sequencing of broad-range gene targets for bacteria and fungi have emerged as important tools to diagnose infections. During the past decade, clinical laboratories have applied PCR amplification and gene sequencing to characterize organisms from culture, and occasionally, to directly detect pathogens from patient samples. Gene sequencing is a more accurate and reproducible method to identify organisms and has increased our ability to capture the diversity of microbial taxa. This is a new technology that has resulted in the identification of unusual organisms and the detection of novel, difficult-to-cultivate organisms, such as Tropherymawhipplei, (.Jung et al., (2019).
However, clinicians are now faced with interpreting microbiological reports that include taxa that are unfamiliar and cannot be assimilated in a meaningful clinical setting. Also, application of broad-range bacterial and fungal PCR directly from clinical material is now more widely available, shifting from research to the clinical setting. This review defines sequence-based identification of bacteria and fungi, with particular emphasis on improving the understanding of increasingly complex world of microbial taxonomy as it relates to the clinical context. Additionally, this review discusses a rational approach to broad-range bacterial PCR and gene sequencing when applied directly to clinical samples, (Guha et al. 2018).
Cheng and Prayogo, (2014) have presented the Symbiotic Organism Search Algorithm, which is an optimization metaheuristic inspired by the symbiotic relationships that occur along with the organisms in nature. In the last few years, the SOS algorithm attracted increasing attention due to its good performance on various real-world problems, despite the fact that no specific parameter adjustment is required. In this paper, we propose an improved version of Symbiotic Organism Search by modifying the organism’s selection strategy. In the offered version of the algorithm, three organisms are selected from the population without having a predefined symbiotic relationship. Once the organisms are selected, an assignment step is conducted to assign each organism to a symbiotic relationship. The testing process was done to analyze the performance of the proposed algorithm using twenty benchmark instances of the flow shop scheduling problem.
The proposed modification improved the performance of the SOS algorithm in the search for the global optimum value in most of the instances. Guha et al. (2018) have shown that user data are aligned, gene information is recognized, and genome structures are compared based on user-defined GenBank files. Information regarding inversion, gain, loss, duplication and gene rearrangement among multiple organisms being compared is provided by geneCo, which uses a web-based interface that users can easily access without any need to consider the computational environment.
Recently, Jung et al., (2019) demonstrated the density of marine organisms, size and direction of the current, an early warning model of marine organism invasion based on BP neural network was established. A quantitative evaluation of the intensity of marine organism invasion can be obtained through the designed early warning model. BPNN is used to estimate the relationship between marine organisms density and invasion intensity in different current velocity (Kanimozhi et al., 2016, Guha et al 2018).
Rodrigues et al (2018) proposed by the author, shows an attempt has been made to incorporate a nature inspired optimization algorithm namely symbiotic organism search (SOS) for optimistic results of load frequency control problem. SOS mimics the symbiotic relations received by an organism to stay in the ecosystem.
Similarly Yang et al., (2018) proposed control technique while they evaluated a two-area multi-unit multi-source power plant equipped with classical controllers. The power system nonlinearities such as generation rate constraint, governor dead band and time delay of transmission system are considered in the study to appraise feasibility of SOS algorithm. The controller settings are concurrently optimized using SOS algorithm via minimization of integral time square error based fitness function. The tuning ability of SOS algorithm is demonstrated by relative study with other optimization techniques reported before. Furthermore, a frequency stabilizer is included in the LFC loop to provide faster damping to the system oscillations. Simulation results shows that proposed coordinated damping controller exhibits greater dynamic performance in terms of overshoot, peak time, settling time (Prakash and Rajathy 2015 ).
Salwa (2019), presented DNA polymorphisms in DNA sequences among individuals, groups, or populations. Polymorphism at the DNA level includes a wide range of variations from single base pair change, many base pairs, and repeated sequences. Genomic variability can be present in many forms, including single nucleotide polymorphisms (SNPs), variable number of tandem repeats (VNTRs, e.g., mini- and microsatellites), transposable elements (e.g., Alu repeats), structural alterations, and copy number variations. Different forms of DNA polymorphisms can be tracked using a variety of techniques; some of these techniques include restriction fragment length polymorphisms (RFLPs) with Southern blots, polymerase chain reactions (PCRs), hybridization techniques using DNA microarray chips, and genome sequencing. During the last years, the recent advance of molecular technologies revealed new discoveries of DNA polymorphisms, (Guha etal 2018).
Taraswinee et al., (2015) have proposed an efficient and reliable Symbiotic Organism Search algorithm for solving Economic Load Dispatch problem. The superiority of Symbiotic Organism Search is revealed for 6 bus system including the transmission limitation. It has been gathered that Symbiotic Organism Search method gives considerable results for Security Constrained Economic Dispatch problem.
Problem Definition: Most commonly existing method is to handle large numbers of biological elements in high-throughput data like microarray data, various gene set-based methods have been proposed with successful applications, and so on. A key idea of the gene set based methods is to evaluate enrichment of the significant genes in the prescribed gene set; this leads to the results biologically more interpretable. All details are maintained manually. The existing systems don’t have the ability to identify these organism facilities. We can only identify several regions.
- Identify several regions only performed.
- Not suitable for all regions.
Proposed System: In the proposed system, user can choose and select any type of organism’s image such asplant, animal, etc.After successful selection the proposed tool automatically analysis the training data set and it finds the name of the organisms along with this, it identifies the specific region with some other related information in an effective manner. This project reduces the manual activity analyzing the details. Using this software tool easily know all the related organisms specific region information also can make report effectively. This makes the system user-friendly thus reducing the manual work.
- Easily identify plant or animal region.
- Easy identification.
Process Involved In Proposed Work Authentication: This module is mainly based on admin. System will check the admin user name and password for authentication. After the verification for authorization the admin can be able to precede the process. All works are done under his control.
Organism and region upload: In this module the admin have to login this application and using their username and password. After successful login admin can upload the Organism and region details in this application. The collection details can be uploaded in this module.
User registration and Login: This module is based on the user control. The user can register their details in the module. After registration, the user can login to the application and view the organism and region details.
Upload image process: In this module used to the analyze region. In this module user upload organism details such as a plant, animal or a single-celled life form, or something that has interdependent parts and that is being compared to a living creature. An example of an organism is a dog, person or bacteria.
RESULTS AND DISCUSSION
This section describes the implementation result and report process. Implementation is the realization of an application, or execution of plan, idea, model, design of a research. This section clarifies the software, datasets and process which are used to develop the research. The proposed system has been successfully implemented which has the effective searching properties and functions.
Software Specifications:The system has used Visual Studio.Net framework. And C#.Net has been used for developing the front end and SQL Server for the back end. The reason for using C#.Net is its flexibility. For the experiment, An Intel I3 2.2 GHz processor with 2 Gb RAM is used to measure the execution time and detection speed.
The table 4.1 shows the proposed software specification of the proposed application development. The main reason of using .Net framework is, it’s a complete GUI and have many unique features to deploy a high featured windows application.
Table 4.1 software specification
| Operating System | Windows 10 |
| Front End | ASP.Net , C# |
| Back End | MS SQL Server |
4.2 Screen Shot
It is the initial form for identification of specific region in organism tool. This tool consists of two login, one is Admin login and another one is user login. Which has shown below :
Home Page:
Figure 1
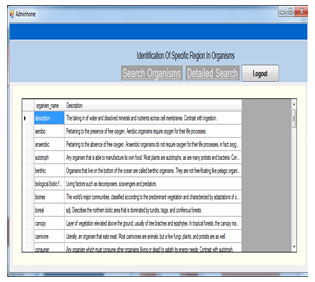
Admin Home: This is the admin home page. The admin process is shown in this form. This page comprises the details like organism and region upload, Upload images, show datas.
Figure 2
Upload Data:This is the page where admin clicks the upload data tab and upload the organism dataset in the page.
Figure 3
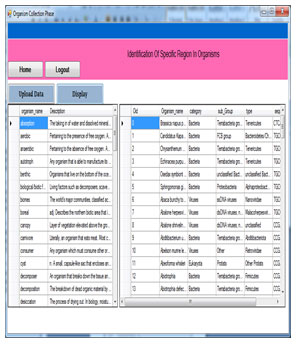
This page shows the uploaded dataset details like organism name and organism description.
Figure 4
Search Organism:This form refers the detailed search for the organism, if the admin clicks the organism tab; this tool displays the detailed description of an organism.
Figure 5
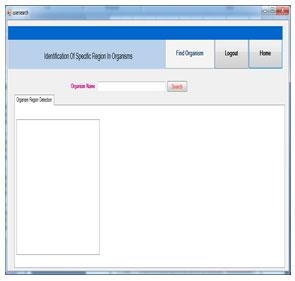
This page isused to search organism. The user can enter organism name and search the organism details.
Figure 6
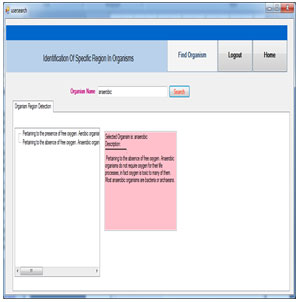
Find Organism:This page shows the region detection details of organism. The Selected organism name and the region descriptions are displayed.
Figure 7
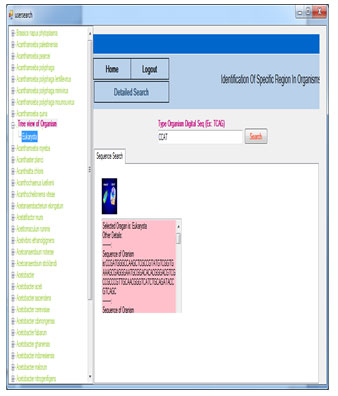
If the user enters sequence id and search the details, it will show the organism name and its descriptions.
Figure 8
CONCLUSION
It is concluded that the application works well and satisfies both the admin and user. The application is tested very well and errors are properly debugged.System can solve the problems from the literature review and it additionally improves the identification of specific regions in the organism. The experimental results are evaluated by using the DotNet environmental area. In this experiment, it shows that a proposed software tool indicates better identification quality assessment compared to the existing models.
REFERENCES
Cheng, M. Y., &Prayogo, D. (2014). Symbiotic organisms search: a new metaheuristic optimization algorithm. Computers & Structures, 139, 98-112.http://dx.doi.org/10.1016/j.compstruc.2014.03.007
Guha D, Provas Kumar Roy, Subrata Banerjee,(2018) Symbiotic Organism Search Based Load Frequency Control with TCSC”, IEEE, 978-1-5386-3039-6
Jung, J., Kim, J. I., & Yi, G. (2019).geneCo: a visualized comparative genomic method to analyze multiple genome structures. Bioinformatics.https://doi.org/10.1093/bioinformatics/btz596
Kanimozhi, G., Rajathy, R., & Kumar, H. (2016). Minimizing energy of point charges on a sphere using symbiotic organisms search algorithm. International Journal on Electrical Engineering and Informatics, 8(1), 29-44.
Prakash, S, &Rajathy, R. (2015). Implementation of symbiotic organism search algorithm for extracting maximum power from the pv system under partially shaded condition. International Journal of Control Theory and Application, 8(5), 1871-1880.
Rajathy, R., Taraswinee, B., & Suganya, S. (2015, March). A novel method of using symbiotic organism search algorithm in solving security-constrained economic dispatch.In 2015 International Conference on Circuits, Power and Computing Technologies [ICCPCT-2015] (pp. 1-8).IEEE.
Rodrigues, L. R., Gomes, J. P. P., Neto, A. R. R., & Souza, A. H. (2018, July). A Modified Symbiotic Organisms Search Algorithm Applied to Flow Shop Scheduling Problems. In 2018 IEEE Congress on Evolutionary Computation (CEC) (pp. 1-7).IEEE.
Salwa Teama (2019), DNA Polymorphisms: DNA-Based Molecular Markers and Their Application in Medicine Genetic Diversity and Disease Susceptibility.
Yang, L., Humin, Z., & Wei, M. (2018). Early warning model for marine organism detection based on BP neural network. In 2018 37th Chinese Control Conference (CCC) (pp. 1909-1914).IEEE.