1Department of Biotechnology, Sri Dharmasthala Manjunatheshwara
College, Ujire, Karnataka, India.
2Department of Pharmacology, JSS Medical College, JSS Academy of Higher
Education and Research, Mysuru, Karnataka, India.
3Department of Biotechnology and Bioinformatics, School of Life Sciences, JSS
Academy of Higher Education and Research, Mysuru, Karnataka, India.
Corresponding author email: prarthanarao6@gmail.com
Article Publishing History
Received: 25/12/2021
Accepted After Revision: 28/02/2022
Amongst the cytoskeletal proteins of bacteria, MreB is known to have very crucial role in modulating shape of the bacteria. Present study involves the use of biocide (A-22) which minimizes the bacterial size augmenting with minimal antibiotic consumption. Intended experiment is designed to be carried out on selected pure strains of gram-positive and gram-negative bacteria namely Lactobacillus rhamnosous ATCC 7469 and Pseudomonas aeruginosa ATCC 27853 respectively. The pure strains are exposed to biocide and changes in the shape is recorded by means of Foldscope (Origami based paper microscope, Prakash Labs) and in-vivo assessment done using antibiotic sensitivity assays with different antibiotics.The novel biocide specifically targeting bacterial cytoskeletal protein, that determines rod shape among bacterial population.
The said compound is also experimented as combinational drug along with conventional antibiotics to reduce antibiotic dose needed to kill and to overcome antibiotic resistance. The A-22has reduced nearly 60-70% antibiotic usage. In Pseudomonas aeruginosa ATCC 27853 when tested for MIC using A-22 and different antibiotics, it was found that 0.5 µg/ml of ampicillin, 1 µg/ml of streptomycin and 5 µg/ml erythromycin were effective in curtailing bacteria against conventional antibiotic concentrations ampicillin 128 µg/ml streptomycin 32 µg/ml, erythromycin 64 µg/ml. Compared to doses of antibiotics required to kill bacteria, the combinational drug of biocide and antibiotic have shown promising effects in killing bacteria at very less concentration, this can useful for treating most diseases caused by antimicrobial resistance bacterial populations.
Antibiotic Sensitivity, Biocide, Cytoskeletal Proteins, Foldscope, Mreb
Prarthana J, Nayana B, Jayanthi M. K, Akhila C, Patil S. M, Ramu R. Biological and Computational Approach to Modify Bacterial Size and Reduce its Antibiotic Consumption Targeting Mreb Bacterial Cytoskeletal Protein. Biosc.Biotech.Res.Comm. 2022;15(1).
Prarthana J, Nayana B, Jayanthi M. K, Akhila C, Patil S. M, Ramu R. Biological and Computational Approach to Modify Bacterial Size and Reduce its Antibiotic Consumption Targeting Mreb Bacterial Cytoskeletal Protein. Biosc.Biotech.Res.Comm. 2022;15(1). Available from: <a href=”https://bit.ly/3CmQDT3“>https://bit.ly/3CmQDT3</a>
Copyright © This is an Open Access Article distributed under the Terms of the Creative Commons Attribution License (CC-BY). https://creativecommons.org/licenses/by/4.0/, which permits unrestricted use distribution and reproduction in any medium, provided the original author and sources are credited.
INTRODUCTION
Eukaryotes and prokaryotes contain cytoskeletal protein structures that are essential to stabilize cell membrane and also provide rigidity to the cell (Soufo and Graumann 2007; Vatset al. 2009).In prokaryotes ithelpsin maintaining cell morphology, cell growth, cell division and chromosome segregation. The cytoskeletal proteins of eukaryotes namely actin, tubulin, and intermediate filaments are homologous to cytoskeletal proteins of bacteria MreB, FtsZ, and crescentin respectively (Daisuke et al. 2008). Among these,MreB protein determines rod-shape in bacteria:MreB which is an actin homolog is recognized as significant protein in maintaining rod shape in the bacteria such as Pseudomonas aeruginosa, Escherichia coli, Caulobacter and Bacillus subtilis (Busieket al. 2015; Awunii 2020).
It was first identified in Escherichia coli as a protein necessary in maintaining cell shape (Doiet al. 1988). MreB belongs to the superfamily of HSP70–actin–sugar kinaseand it was known in forming spirals which can traverse the longitudinal axis of cells of B. subtilis, this suggests that bacteria are having an internal actin-like cytoskeleton to maintain cell shape which isanalogous to eukaryotes (Joneset al. 2001; Awuni and Mu 2019).
It is considered to be conserved actin homolog in prokaryotes and is mainly encoded in the chromosomes of the bacterial species and also helps in variety of cellular activities. A22 – biocide changing rods into cocci:S-(3,4-dichlorobenzyl)isothiourea (A22) biocide is a derivative of S-benzylisothiourea which is chemically designated as [(3,4-dichlorophenyl)methyl]thiocarboxamidine (Figure1) (Iwaiet al. 2002; Carballido-Lopez et al. 2006; Awunii 2020).
Figure 1: Structure of A-22
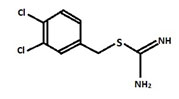
The A22 is known to be acting like a reversible inhibitor of a bacterial cell wall protein MreB which leadsto change in shape of bacteria from rods to the coccoid form and also it preventsassembly of MreB into long rigid polymers. As a result of change in shape various properties of the bacteria can be affected, such as the cell division, the acquisition of nutrients, motility, the clamping surfaces, and pathogenesis (Bonezet al. 2016).
To check the activity of A22 in changing rods to cocci the pure strains were used in this study which includes Pseudomonas aeruginosa ATCC 27853, Escherichia coli ATCC 25922, Lactobacillus rhamnosus ATCC 7469 and Lactobacillus casei ATCC 393 (Valik et al. 2008; Percival and Williams 2014; Moradali et al. 2016; Awuni 2020).
P. aeruginosa and E. coli were used in the study as they are pathogenic microorganisms and they show resistance to most of the antibiotics. As a case combination of biocide and antibiotics are used to inhibit the bacterial growth.In the current scenario, several phytochemical formulations have been used to treat microbial diseases (Patil et al. 2021a; Patil et al. 2021b; Patil et al. 2021c). Small molecules like bioactive peptides have also been used, indicating the significance of small chemical compounds (Patil et al. 2020).
In the present study MreB polymerization is targeted using A22, MreB is very essential for cell wall biosynthesis and is also conserved in all rod shaped bacteria. This study mainly involves the use of combination of biocide and antibiotics against strains such as P. aeruginosa and E. coli because of their resistance to almost all the antibiotics. Their size reduction is established with minimum amount of conventional antibiotic to kill them. By this treatment of many severe diseases caused by these bacteria can be made possible.
MATERIAL AND METHODS
The pure strains of the bacteria from ATCC were taken and streaked on the agar plates. Pseudomonas aeruginosaand Escherichia coliwas grown on Nutrient agar (NA) and incubated at 37°C for 24 hrs whereas, Lactobacillus rhamnosus was grown on De Man, Rogosa and Sharpe (MRS) agar and incubated at 37°C for 24 hrs (Bonez et al. 2016).Different broth was used for inoculating different bacteria.
For P. aeruginosa Mueller Hinton borth (MHB) and agar was used for inoculating and plating respectively, whereas for E. coli Luria bertani broth and agar (LBA) was used for inoculation and plating respectively. L. rhamnosus was inoculated and plated on MRS media. Initially the cultures were exposed to different concentration of biocide (A22) given in the Table 1and incubated at 37°C for 24 hrs.
After 24 hrs, the optical density of the culture was measured at 660nm using UV-VIS Double Beam Spectrophotometer 2205 (Systronics) and the size of the bacteria was measured using micrometry shown in the Table 2 (Awuni and Mu 2019). Then the lawn culture was prepared using sterile swab and different concentration of antibiotics was added and plates were incubated at 37°C for 24 hrsand the zone of inhibition was measured which is given in the Table 3.The bacteria were inoculated with combination of biocide and antibiotics and after 24 hrs of exposure the optical density of the culture was measured using UV-VIS Double Beam Spectrophotometer 2205 (Systronics), and the graph obtained is given in the Figures below (Al-Khayyat et al. 2019).
Molecular docking simulation is performed to understand the interaction of ligands with the target proteins at the molecular level. In this study, the crystallographic structure of MerB protein was retrieved from the RCSB PDB database (PDB ID: 1JCE).
On the other hand, chemical structure of the compound A-22 was drawn and 3D optimized using ChemSketch. Protein and ligand preparation, as well as binding site prediction steps were completed according to the previous studies conducted by Patil et al. (2021d) and Patil et al. (2021e). AutoDock Vina 1.2, an open-source command line software designed for the docking of the molecular entities was used for the docking simulation (Trot and Olson 2010).
The visualization of docking simulation was done using BIOVIA Discovery Studios Visualizer (2021), an open source visualizing GUI software. Druglikeliness and pharmacokinetic analysis, also known as ADMET (adsorption, distribution, metabolism, excretion, and toxicity) predictions, were performed to assess its oral bioavailability of potential drug candidates in silico. In this study, chemical structure of the compound A-22 in SMILES format was submitted to the ADMETlab server (https://admetmesh.scbdd.com/).
For the druglikeliness evaluation, Lipinski’s rule of five was considered. For pharmacokinetic evaluation, CACO-2 permeability, human intestinal absorption (HIA), volume distribution (VD), cytochrome P (CYP) inhibition, hERG blocking, and AMES toxicity parameters were considered (Patil et al. 2021f). As the druglikeliness and pharmacokinetic studies were performed in silico, there were no ethical clearance, patient consent, or any kind of approvals required.
RESULTS AND DISCUSSION
The different bacterial culturesviz.,P. aeruginosa, E. coliand L. rhamnosuswere exposed with 1, 3 and 0.5 µg/ml concentration of the biocide. The optical density of the culture after exposure with biocide and control (without biocide) was recorded and also the size of the bacteria measured is given in the Table 1. After the cultures were exposed to biocide, they were subjected to antibiotic assay against different antibiotics and the zone of inhibition formed is given in the Table 2.The optical density of the culture was recorded and the graph obtained is shown below (Figure 2, 3, and 4). The MIC of the experimental molecules has been shown in Figure 5.
Table 1. Optical density and the size of the bacteria after exposure to A-22 biocide
| Bacteria | Biocide concentration | Optical density (660nm) | Size (µm) |
| Pseudomonas aeruginosa | Control | 0.612 | 7.5 |
| 1 µg/ml | 0.410 | 5.0 | |
| Escherichia coli | Control | 1.453 | 2.5 |
| 3 µg/ml | 0.751 | 1.875 | |
|
Lactobacillus rhamnosus |
Control | 1.684 | 8.0 |
| 0.5 µg/ml | 1.571 | 7.5 |
Table 2. Zone of inhibition formed using different concentration of different antibiotics
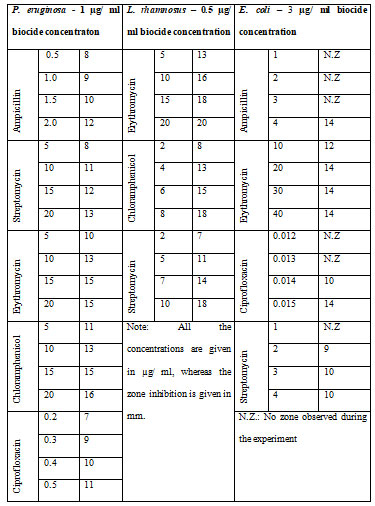
Figure 2: Graphs showing growth percentage of Pseudomonas aeruginosaagainst different experimental
compounds A) ampicillin, B) streptomycin, C) erythromycin, D) chloramphenicol, and E) ciprofloxacin
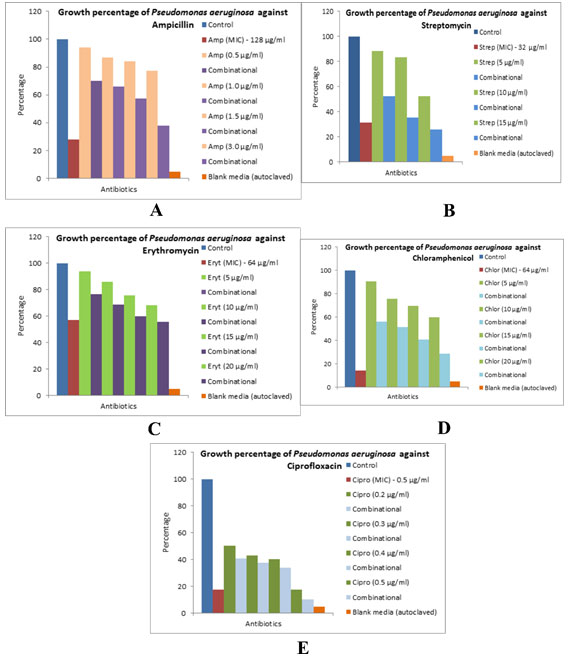
Figure 3: Graphs showing growth percentage of Lactobacillus rhamnosus against different experimental
compounds A) erythromycin, B) streptomycin, and C) chloramphenicol
Figure 4: Graphs showing growth percentage of Escherichia coli against different experimental
compounds A) ampicillin, B) streptomycin, C) erythromycin, D) ciprofloxacin
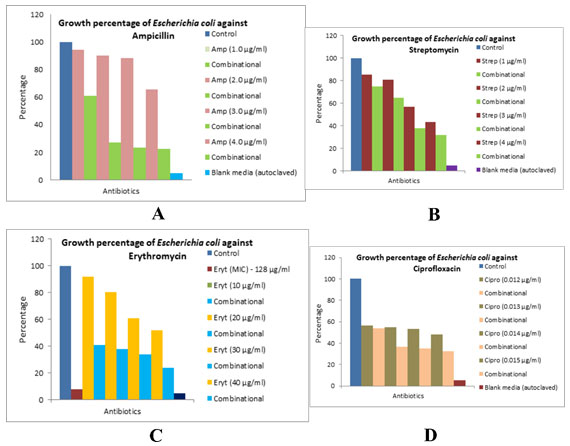
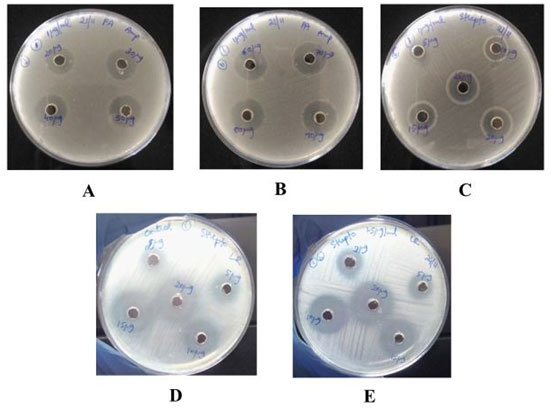
Figure 5: Image of Pseudomonas aeruginosa culture showing zone of inhibition for different concentration of A and B) ampicillin,C) streptomycin, D) Image of Lactobacillus rhamnosus culture showing zone of inhibition (diffuse growth present)
for different concentration of streptomycin without exposing to biocide, E) after biocide exposure
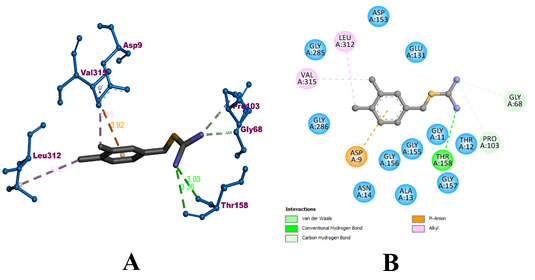
During the molecular docking process compound A-22 formed 3 hydrogens with GLY 68, PRO 103, and THR 158 residues of the protein. It also formed 2 hydrophobic alkyl bonds with LEU 312 and VAL 315. In addition, an electrostatic pi-anion bond with ASP 9 of the protein. The compound was predicted with a binding affinity of -8.7 kcal/mol. The binding interaction of A-22 with MerB has been visualized in Figure 6. The bound amino acids have been colorized according to their binding type. Whereas, the surrounding amino acids have been colored in teal.
Figure 6: Visualization of binding interaction of the compound A-22 with MerB protein in A) 3D and B) 2D
During the evaluation of druglikeliness according to the Lipinski rule, the molecular weight (MW) of A-22 was found to be 227.07 g/mol, which has not exceeded the limit of 500 g/mol. clogP value represents the partition coefficient between n-octanol and water to measure the hydrophilicity. Low hydrophilicities and therefore high clogP values cause poor absorption or permeation. In this study, the clogP was found to be 1.65. Hydrogen bond acceptors were predicted to be 2, whereas the limit has been set for 10. In case of hydrogen bond donors, A-22 was predicted to have 2, with the limit set for 5. Therefore, compound A-22 never violated the rule of Lipinski rule.
In case of pharmacokinetic analysis, Caco-2 cell line permeability was predicted to be -4.97 for A-22. The limit of Caco-2 value is -5.15 log units. Human intestinal absorption value was predicted to be positive (0.008). Volume distribution was predicted to be 4.628, which should be in the range of 0.04-20L/kg. In case of cytochrome P inhibition, A-22 was predicted with no inhibition of cytochrome P enzymes. The human Ether-à-go–Related Gene (hERG) is blocking was predicted with positive value (0.032). Furthermore, AMES toxicity value was predicted to be positive (0.236), which indicated absence of risk of toxicity.
Table 3 represents the druglikeliness and pharmacokinetic analysis of the compound A-22.
Figure 7 visualizes the druglikeliness and pharmacokinetic analysis of A-22.
Figure 7: Visualization of druglikeliness and pharmacokinetic analysis of the A-22. Presence of
blue line represents the compound A-22 within the boundaries of all the parameters used.
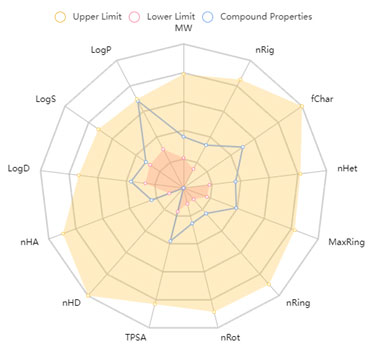
Table 3. Druglikeliness and pharmacokinetic analysis of compound A-22.
| Categories | Types of parameters | A-22 |
| Druglikeliness based on Lipinski’s rule of five | Molecular weight | 227.07 g/mol |
| No. of hydrogen bond donors | 2 | |
| No. of hydrogen bond acceptors | 2 | |
| cLog P | 1.65 | |
| Adsorption | Caco-2 permeability | -4.97 |
| Human intestinal absorption (HIA) | 0.008 | |
| Metabolism | CYP1A2 inhibition | No |
| CYP2C19 inhibition | No | |
| CYP2C9 inhibition | No | |
| CYP2D6 inhibition | No | |
| CYP3A4 inhibition | No | |
| hERG blocking | Clearance (CL) | 0.032 |
| Toxicity | AMES toxicity | 0.236 |
| Distribution | Volume distribution (VD) | 0.071 |
The alarming increase in antibiotic resistance is due poor public health, inexpensive antibiotics which is causing threats in neonatal sepsis, causing therapeutic failures in bacterial infections (Laxminarayan et al. 2015). It is alarming that although bacterial resistance continues to emerge, the rate at which antibiotics are being developed is decreasing (Pulcini et al. 2012). MreB is a promising drug target because it is conserved and essential in most rod-shaped bacteria, MreB has been associated with essential subcellular processes including cell wall biosynthesis and maintenance of cell shape (Doi et al. 1988; Bean and Amann 2008;Awuni 2020).
Determinations of the DNA-sequence of the MreB-gene and of the gene-products of the Mre-region that function in formation of the rod shape of Escherichia coli cells, cell division, cell wall morphogenesisMreB have been identified as potential targets for antibiotics. The nucleotide binding site is an important target for antibiotics development because nucleotide binding plays a crucial role in the structure and dynamics of MreB (Wachi and Matsuhashi 1989; Jones et al. 2001; Soufo and Graumann 2005; Bean and Amann 2008; Awuni and Mu 2019).
ATP induces the polymerization of MreB into filaments required for cell wall biosynthesis (. Interestingly, the polymerization of MreB induces ATP hydrolysis, which serves as a timing process to coordinate depolymerization (Bean and Amann 2008; Gunning et al. 2015). Thus, ATP is required by MreB to function properly and any molecule that could compete with ATP for binding to the nucleotide binding pocket could be a bactericidal agent (Awuni 2020).
The results obtained from this study are the first to evaluate the effectiveness of biocide A22 by inhibiting cytoskeletal protein Mreb on the strains Pseudomonas aeruginosa ATCC 27853 and Lactobacillus rhamnosus ATCC 7469 and to reduce consumption of antibiotics due to decreased size. The MIC value of A22 on Pseudomonasaeruginosa was found be lower in this study, compared to the MIC reported in previous studies (Bonez et al. 2016).
The drastic reduction in the antibiotic consumption, below MIC after the exposure of the strains to biocide was observed in this study. The result reported in the study provides an alternative method to inhibit multi-drug resistant (MDR) microorganisms. The results obtained helps indicates that the biocide A22 used in the study brought about change in bacterial conformation by targeting its cytoskeletal protein MreB, and also reduced antibiotic consumption of bacteria (Awuni 2020).
Apart from the in vitro evaluations, in silico studies conducted on MreB inhibition also indicates that compound A-22 has the higher inhibitory potential. A recent study showed that MreB protein can be inhibited by few of the 100 natural compounds tested. Apart from amentoflavone and rutin, the other compounds failed to achieve significant inhibition. The compounds were also reported with insignificant druglikeliness and pharmacokinetics results (Al-Khayyat et al. 2019).
In another study, phytochemicals from Leucas aspera were screened for the inhibition of MerB in silico. Among them, leucasperone B and penicillin were found to be the potent inhibitors of the protein. However, compound A-22 has been proved with the better outcomes during in silico analysis in comparison with these studies, with respect to binding interaction, druglikeliness and pharmacokinetics analysis. From these we can say that A22 can be used as a novel drug for treating diseases caused by MDR Pseudomonas aeruginosa and also usage of high dose antibiotics can be stopped which can prevent many side effects caused by antibiotics to humans(Sharavanan et al. 2019; Awuni 2020).
CONCLUSION
The findings of this study are the first to assess the efficacy of biocide A22 in suppressing the cytoskeletal protein MreB of Pseudomonas aeruginosa ATCC 27853 and Lactobacillus rhamnosus ATCC 7469, as well as the reduction in antibiotic consumption due to reduced size. This study discovered a significant decrease in antibiotic consumption below the MIC level and there is nearly 60-70 percent antibiotic usage after the strains were exposed to biocide. In addition, our computational investigation also suggests that biocide A-22 inactivates MreB. During druglikeliness and pharmacokinetics analysis reveals A-22 shows no toxic effects. Therefore, we conclude biocide A-22 as a potent anti-bacterial agent against MDR bacterial species.
ACKNOWLEDGEMENTS
This study was financially supported by the DBT (Sanction Order No.BT/IN/Indo-US/Foldscope/39/2015) in partnership with Foldscope Instruments Inc. and Prakash lab (Stanford University; California) from 2018-2019.
Conflict of Interests: Authors declare no conflicts of interests to disclose.
Data Availability Statement: The database generated and /or analysed during the current study are not publicly available due to privacy, but are available from the corresponding author on reasonable request.
REFERENCES
Al-Khayyat, M. Z., Ameen, A. G., and Abdulla, Y. A. (2019). Virtual Screening for Inhibitors Targeting the Rod Shape-Determining Protein in Escherichia coli. Jordan J Biol Sci 12(2).
Awuni, E., and Mu, Y. (2019). Effect of A22 on the conformation of bacterial actin MreB. Int J Mol Sci 20:1304.
Bean, G. J., and Amann, K. J. (2008). Polymerization properties of the Thermotoga maritima actin MreB: roles of temperature, nucleotides, and ions. Biochemistry 47, 826–835.),
Bonez P. C., Ramos, A. P., Nascimento K., et al. (2016). Antibacterial, cyto and genotoxic activities of A22 compound ((S-3,4-dichlorobenzyl) isothiourea hydrochloride). Microb Pathog 99 14–18.
Bratton, B.P., Shaevitz, J.W., Gitai, Z. et al. (2018). MreB polymers and curvature localization are enhanced by RodZ and predict E. coli’s cylindrical uniformity. Nat Commun 9(1), 1-11.
Busiek, K. K., and Margolin, W. (2015). Bacterial actin and tubulin homologs in cell growth and division. Curr Biol 25, R243–R254.
Carballido-López, R., Formstone, A., Li, Y., et al. (2006). Actin homolog MreBH governs cell morphogenesis by localization of the cell wall hydrolase LytE. Dev Cell. 11(3); 399-409
Daisuke, S., Masako, S., andHironori, N. (2008). Determination of bacterial rod shape by a novel cytoskeletal membrane protein. EMBO J 27(23):3081-91
Doi, M., Wachi, M., Ishino, F., et al. (1988). Determinants of the DNA sequence of the mreB gene and of the gene products of the mre region that function in formation of the rod shape of Escherichia coli. J Bacteriol 170, 4619–4624.
Awunii, E., (2020). Status of targeting MreB for the development of antibiotics. Front Chem. http://doi.org/10.3389/fchem.2019.00884
Figge, R. M., Divakaruni, A. V., and Gober, J. W. (2004). MreB, the cell shape-determining bacterial actin homologue, co-ordinates cell wall morphogenesis in Caulobacter crescentus. Mol Microbiol 51(5), 1321–1332.
Gunning, P. W., Ghoshdastider, U., Whitaker, S., et al. (2015). The evolution of compositionally and functionally distinct actin filaments. J Cell Sci 128, 2009–2019.
Harold, P. E.(2017). The discovery of the prokaryotic cytoskeleton: 25th anniversary. Mol Biol Cell. 28(3): 357–358.
Iwai N., Nagai K., and Wachi M. (2002). Novel S-benzylisothiourea compound that induces spherical cells in Escherichia coli probably by acting on a rod-shape-determining protein(s) other than penicillin-binding protein 2. Biosci Biotechnol Biochem 66:2658–2662.
Jones, L. J. F., Carballido-López, R., and Errington, J. (2001). Control of cell shape in bacteria: helical, actin-like filaments in Bacillus subtilis. Cell 104, 913–922.
Laxminarayan, R., Matsoso, P., Pant, S., et al. (2015). Access to effective antimicrobials: a worldwide challenge. Lancet 387: 168–175
Lim, J.Y., Yoon, J.W. and Hovde, C.J., (2010). A Brief Overview of Escherichia coli O157:H7 and Its Plasmid O157. J Microbiol Biotechnol. 20(1): 5–14.
Moradali, M.F., Ghods, S. and Rehm, B.H. (2017). Pseudomonas aeruginosa lifestyle: a paradigm for adaptation, survival, and persistence. Front Cellular Infect Microbiol 7, 39.
Patil, S. M., Kumari, V. C., Sumana, K., et al. (2021c). Sustainable development of plant tissue culture industry: The Indian scenario. J Appl Biol Biotechnol 9(2): 18-27.
Patil, S. M., Maruthi, K. R., Naik, S., et al. (2021f). Comparative molecular docking and simulation analysis of molnupiravir and remdesivir with SARS- CoV-2 RNA dependent RNA polymerase (RdRp). Bioinformation 17(11); 932-933.
Patil, S. M., Ramu, R., Shirahatti, P. S., et al. (2021b). A systematic review on ethnopharmacology, phytochemistry and pharmacological aspects of Thymus vulgaris Linn. Heliyon, 7(5):e07054.
Patil, S. M., Shirahatti, P. S., and Ramu, R. (2021a). Azadirachta indica A. Juss (neem) against diabetes mellitus: a critical review on its phytochemistry, pharmacology, and toxicology. J Pharm Pharmacol https://doi.org/10.1093/jpp/rgab098
Patil, S. M., Sujay, S., Tejaswini, M., et al. (2020). Bioactive peptides: its production and potential role on health. Int J Innov Sci Eng Technol. (7) 167-182.
Patil, S.M., Martiz, R.M., Ramu, R., et al. (2021d). Evaluation of flavonoids from banana pseudostem and flower (quercetin and catechin) as potent inhibitors of α-glucosidase: an in silico perspective. J Biomol Struct Dyn. 1-15. DOI: 10.1080/07391102.2021.1971561
Patil, S.M., Martiz, R.M., Ramu, R., et al. (2021e). In silico identification of novel benzophenone-coumarin derivatives as SARS-CoV-2 RNA dependent RNA polymerase (RdRp) inhibitors. J Biomol Struct Dyn. 1-17. DOI: 10.1080/07391102.2021.1971598
Percival, S., and Williams, D. (2014). Escherichia coli.In Microbiology of Waterborne Diseases. Academic Press USA (2) Pages 89-117.
Peter, L.G.,(2007). Cytoskeletal elements in bacteria. Annu Rev Microbiol. 61:589-618.
Pulcini, C., Bush, K., Craig, W.A., et al. (2012). Forgotten antibiotics: an inventory in Europe, the United States, Canada, and Australia. Clin Infect Dis 54, 268–274.
Sharavanan, V.J., Sivaramakrishnan, M., Kothandan, R., et al. (2019) Molecular docking studies of phytochemicals from Leucas aspera targeting Escherichia coli and Bacillus subtilis subcellular proteins. Pharmacogn J. 11(2).
Soufo, H. J. D., and Graumann, P. L. (2005). Bacillus subtilis actin-like protein MreB influences the positioning of the replication machinery and requires membrane proteins MreC/D and other actin-like proteins for proper localization. BMC Cell Biol. 6:10.
Trott, O. and Olson, A.J. (2010). AutoDock Vina: improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J Comput Chem 31(2):455-61.
Valík, L., Medvedová, A., Čižniar, M., et al. (2008). Characterization of the growth of Lactobacillus rhamnosus GG in milk at suboptimal temperatures. J Food Nutr Res47, 60-67.
Vats, P., Yu, J. and Rothfield, L. (2009). The dynamic nature of the bacterial cytoskeleton. Cell Mol Life Sci. 66(20): 3353–3362.
Wachi, M., and Matsuhashi, M. (1989). Negative control of cell-division by MreB, a gene that functions in determining the rod shape of Escherichia coli cells. J. Bacteriol. 171, 3123–3127.
Weng, Y., Chen, F., Liu, Y., et al. (2016). Pseudomonas aeruginosa Enolase Influences Bacterial Tolerance to Oxidative Stresses and Virulence. Front microbial 7.


