1Department of Biotechnology, K. L. E. Technological University, Hubballi, Karnataka, India.
2Department of Chemical Engineering, K. L.E., Dr. M. S. Sheshgiri College of Engineering
and Technology, Belgaum, Karnataka, India.
Corresponding author email: anilrshet@gmail.com
Article Publishing History
Received: 25/08/2021
Accepted After Revision: 15/11/2021
Due to the increased use of synthetic dyes in various industries, there is an increased disposal of wastewater containing harmful dyes. These, in turn, have affected plants, animals, and humans. The physical and chemical methods of dye decolorization have failed to degrade the synthetic dyes in industrial effluents completely. The microbial decolorization is better due to its versatility, dynamic metabolism, and potential machinery of enzymes. This study aimed to degrade basic yellow dye auramine O by bacteria isolated from textile industry effluent. In this regard, five bacterial strains were isolated and screened from a soil sample taken from textile industry effluent.
The initial physical and biochemical characterization of the bacterial isolates 1 and 2 indicated catalase test-positive, starch test-negative, motility agar test-negative, gram staining test-positive, and morphology-bacillus. The bacterial isolates 3, 4, and 5 indicated oxidase test-negative, urease test-positive, gram staining test-negative, and morphology-staphylococcus. All the isolates were further subjected to a screening test, where isolate 5 showed maximum dye decolorization of 98.9% in 96 h.
The biodegradation of dye was optimized for different values of initial pH (4-10), inoculum size (2% -10%), initial dye concentration (50 mgL-1 to400 mgL-1), carbon source (glucose, fructose, xylose, starch and lactose) and nitrogen source (peptone, ammonium sulphate, yeast extract, ammonium nitrate and urea). Maximum dye decolorization was observed for initial dye concentration of 200 mgL-1, initial pH of 6, inoculum size of 10%, yeast extract as nitrogen source, and glucose as carbon source. Therefore, dye degradation by bacteria can be used as a potential method for auramine O dye treatment.
Auramine O Dye, Bacteria, Biodegradation, Decolorization, Textile Industry.
Shet A. R, Patil L. R, Hombalimath V. S, Achappa S, Desai S. V, Kadapure S. A. Biodegradation of Basic Yellow Auramine O Dye using Staphylococcus spp. Isolated from Textile Industry Effluent. Biosc.Biotech.Res.Comm. 2021;14(4).
Shet A.R, Patil L.R, Hombalimath V.S, Achappa S, Desai S.V, Kadapure S.A. Biodegradation of Basic Yellow Auramine O Dye using Staphylococcus spp. Isolated from Textile Industry Effluent. Biosc.Biotech.Res.Comm. 2021;14(4). Available from: <a href=”https://bit.ly/3HhxVy0“>https://bit.ly/3HhxVy0</a>
Copyright © Shet et al., This is an open access article distributed under the terms of the Creative Commons Attribution License (CC-BY) https://creativecommns.org/licenses/by/4.0/, which permits unrestricted use distribution and reproduction in any medium, provide the original author and source are credited.
INTRODUCTION
Dyes are an essential component in different industries like paper, food, leather, textile, paint, and cosmetics. Roughly around 25 dye groups are present based on the chemical structure of chromophore. Many dyes are grouped as textile dyes which are used to dye different fabrics (Shet et al. 2013; Hombalimath et al. 2012; Shet et al. 2016; Kulkarni et al. 2017; Abe et al. 2019; Benkhaya et al. 2020). Disposal of industrial- effluents to the water bodies causes water pollution. The environmental pollution can cause health risks to all the living things on the earth (Varjani 2017; Varjani and Upasani 2017; Bencheqroun et al. 2019; Shet et al. 2021; Achappa et al. 2021).
The dyes used in textile industry causes artistic damage and stops the light diffusion in water, leading to a decreased level of dissolved oxygen and thus affecting the aquatic life’s photosynthesis rate (Ajaz et al. 2020; Achappa et al. 2020). Among different dyes, Auramine O is a diarylmethane dye that is used as a fluorescent stain in paper, leather, textile, and paint industries. Various methods are used to remove the auramine O dye from textile effluents: physical, chemical, Physico-chemical, and biological (Cao et al. 2019; Varjani and Upasani 2019; Nakkeeran et al. 2020; Achappa et al. 2021; Shet et al. 2021).
The biological method has many advantages like; it is simple, cheap, and environmentally friendly process. The large numbers of microbes present are maintained easily and require less preparation (Kulkarni et al. 2017; Crini and Lichtfouse 2018). Different microbes such as fungi, algae, bacteria, and yeast decolorize different dyes (Roy et al. 2018; Shet et al. 2020; Bhavikatti et al. 2020). Textile wastewater treatment can be done using pure or mixed microbial cultures (Tochhawng et al. 2019; Patil et al. 2020).
Different factors like nutrients, soluble salts, initial pH, heavy metals, incubation temperature, dye structure, etc., affect the dye decolorization (Al-Amrani et al. 2014). There are different reports available which showed different dye decolorization using microbes (Varjani and Upasani 2016; Kiayi et al. 2019; Pratiwi et al. 2019; Achappa et al. 2021; Shet et al. 2021). In the Present study, biodegradation of basic yellow dye auramine O using bacteria has been reported. The effect of inoculums size, initial pH, carbon source, initial dye concentration, nitrogen source, and incubation temperature has been examined and the results obtained are discussed.
MATERIAL AND METHODS
Media components were obtained from Himedia laboratories Pvt. Ltd. All the analytical grade chemicals were purchased from S.R.L. Pvt. Ltd. U.V. Visible Spectrophotometer (Make: Metler Toledo) was used for the analysis of the samples. The samples were collected from different disposal points of the local textile industry in the form of soil samples. Sterile plastic bottles and spatula were used to collect the samples. It was then stored in the laboratory at 4oC. The soil samples were cultured on sterile nutrient agar plates, and 5 isolates were screened and transferred to sterile slants and preserved.
For screening, Bushnell and Hass’s mineral medium was prepared using a standard protocol. The broth pH was adjusted to 7. The dye of concentration 100 mgL-1was added to the test tubes. This was distributed in test tubes and autoclaved to maintain sterility. The isolates were added to separate test tubes and incubated. Initial studies were done to decide on the operating environment such as initial pH (7), substrate concentration (100 mgL-1), and incubation temperature (30oC).
Percentage dye decolorization was determined by taking 1.5 mL aliquots from time to time from different test tubes. Then the sample was centrifuged at 5,000 rpm for 15 minutes to take out the cells. The supernatant was then measured for its absorbance at 370 nm using UV-visible spectrophotometer. The percentage dye decolorization was determined by using the mathematical expression: % Dye decolorization = (C – T/C) × 100. Where C = Initial absorbance, T = Final absorbance.
Standard gram staining and microscopic observations were carried out to characterize the isolates. The isolates were transferred on the slide with a sterile inoculating loop. It was allowed to dry. A drop of violet stain was put on the culture and waited for 30 seconds, then washed under running water carefully. A drop of iodine was added and waited for 30 seconds to allow it to dry. Then safranin stain was added and waited for 30seconds till it dried and washed under running water carefully.
The slide was then observed under a microscope [100X] with immersion oil. Biochemical characterization like oxidase test, catalase test, urease test, starch hydrolysis test, and motility agar test were conducted on the isolates and recorded. Different media and process parameters were optimized by One Factor At a Time (OFAT) to get the highest decolorization efficiency of auramine O dye by the selected bacterial isolates. The experiments were performed in triplicates. The effect of different pH (4, 6, 8, and 10) was observed for the dye decolorization efficiency of the selected bacterial isolate.
The Bushnell and Hass medium was spiked with auramine O dye of concentration 100 mgL-1 and was incubated at a temperature of 30°C. The initial value of pH was set using 0.01 M HCl / 0.01 M NaOH solutions. Percentage dye decolorization was determined. The effect of three different initial dye concentration (50, 200, and 400 mgL-1) of Auramine O dye was examined on the dye decolorization efficiency.
The inoculum was added to 10 mL medium, maintained at 1:50 inoculum to broth ratio and incubation temperature of 30°C. Percentage dye decolorization was determined by taking 1.5 mL aliquots from time to time from different test tubes. The samples were centrifuged at 5,000 rpm for 15 minutes to take out the cells. The supernatants were then measured for its absorbance at 370 nm using UV- visible spectrophotometer, and % decolorization was calculated. Abiotic decolorization was checked using an un-inoculated blank.
Different inoculum sizes (2% and 10%) were used to examine the effect on the dye degrading efficiency of the selected bacterial isolates. The Bushnell and Hass medium was spiked with auramine O dye of concentration 100 mgL-1 and was incubated at a temperature of 30°C.Percentage dye decolorization was determined. To analyze the effect of media components on the bacterial dye degradation, five different carbon sources: fructose, glucose, xylose, starch, and lactose, and five different nitrogen sources: peptone, ammonium sulphate, yeast extract, urea and ammonium nitrate were used (4 gL-1). The Bushnell and Hass medium was spiked with auramine O dye of concentration 100 mgL-1 and was incubated at a temperature of 30°C. Decolorization of the dye was determined.
RESULTS AND DISCUSSION
Physical and Biochemical characterization: The soil samples were collected from local textile industry effluent where auramine O dye was used for dyeing purposes. The organisms were cultured on agar slants, and they were identified by gram staining. The biochemical and microscopic characterization of the bacterial isolates are shown in table 1 and 2 respectively. The biochemical tests and gram staining indicated that the culture was a mixed culture that contained both gram-positive and gram-negative organisms.
Some of them were rod-shaped, and some of them were round-shaped. Bacterial isolates 1 and 2 showed catalase test- positive, starch test- negative, motility agar test- negative, and the gram staining test showed that they were gram-positive strains and bacillus morphology. The bacterial isolates 3, 4 and 5 were staphylococcus as they were gram-negative, oxidase test negative, and urease test positive.
Table 1. Biochemical characterization of the bacterial isolates
| Test | Bacterial Isolates | ||||
| 1 | 2 | 3 | 4 | 5 | |
| Gram staining | (+) ve | (+) ve | (-) ve | (-) ve | (-) ve |
| Catalase test | (+) ve | (+) ve | -NA- | -NA- | -NA- |
| Motility agar | (-) ve | (-) ve | (-) ve | (-) ve | (-) ve |
| Starch agar | (-) ve | (-) ve | -NA- | -NA- | -NA- |
| Urease | -NA- | -NA- | (-) ve | (-) ve | (-) ve |
| Oxidase | -NA- | -NA- | (+) ve | (+) ve | (+) ve |
Table 2. Microscopic characterization of the bacterial isolates
| Property | Bacterial Isolates | ||||
| 1 | 2 | 3 | 4 | 5 | |
| Elevation | Flat | Flat | Flat | Flat | Flat |
| Margin | Irregular | Irregular | Irregular | Irregular | Irregular |
| Opacity | Opaque | Opaque | Opaque | Opaque | Opaque |
| Colour | Pale | Pale | Pale | Pale | Pale |
| Surface | Moist, shiny | Moist, shiny | Moist, shiny | Moist, shiny | Moist, shiny |
| Gram staining | (+) ve | (+) ve | (-) ve | (-) ve | (-) ve |
| Shape of vegetative cell | Rod | Rod | Coccus | Coccus | Coccus |
Biodegradation of Auramine O dye: All the five bacterial isolates showed effective ability in decolorizing auramine O dye. Degradation started in few hours, and after 96 h, the average percentage of dye removal by Bacillus spp. was 96% and by staphylococcus spp. was 98%. No color decrease was observed in a sterile control. At 24 h, isolate 5 showed a higher percentage of dye decolorization when compared to other bacterial isolates. For all the bacterial isolates, the % dye decolorization was observed to increase with time. At 96 h, isolate 5 showed the maximum dye decolorization of 98.9%, as shown in figure 1. Since isolate 5 showed maximum dye decolorization capacity, it was considered for further optimization studies.
Figure 1: Percentage dye decolorization by different bacterial isolates
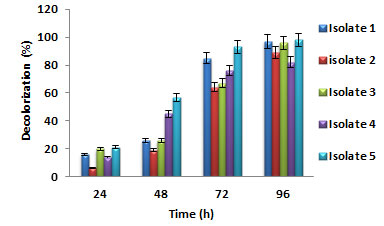
Bacterial oxido-reductive enzymes were responsible for the textile dye decolorization. The bacterial metabolism enabled the use of complex xenobiotic compounds of the dyes and breaks them down into less complex metabolites. The bacteria obtained from actual effluent disposal sites have the enzymes activated, which assisted the decolorization of dye (Radia and Romana, 2019; Ajaz et al. 2020; Shet et al. 2021).
Effect of initial pH: pH is a vital variable for bacterial growth and an important factor for wastewater treatment. Depending on the dye type, pH may be acidic, alkaline, or neutral. Dye decolorization rate may change with the change in pH of the wastewater. Dye decolorization can be improved by modifying the wastewater pH to support bacteria degrading the dye or by choosing the microbes that can grow at the pollutant’s pH (Al-Amrani et al. 2014; Varjani and Upasani 2017).
In this study, as shown in (figure 2), maximum dye decolorization was seen for pH of 6, followed by pH 8. Highest dye decolorization at an initial pH of 8 to 10 using Lysinibacillus boronitolerans CMGS-2 was reported in the previous studies (Basutkar and Shivannavar 2019). Decolorization of malachite green of 98% was obtained by RuO2–TiO2 and Ti mesh electrodes coated with Pt at an initial pH of 4.5 (Singh et al. 2016; Achappa et al. 2021).
Figure 2: Percentage dye decolorization for different initial pH
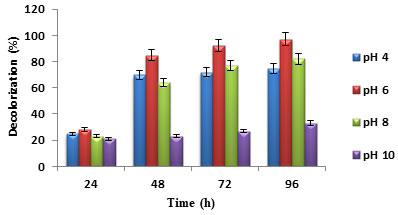
Effect of initial dye concentration: Initial dye concentration affects the dye decolorization. The enzymes produced by dye degrading bacteria may not identify a low concentration of dye. But the high concentration of dye is toxic to bacteria and affects dye decolorization by blocking active sites of enzyme. Similarly, simple structured and low molecular weight dyes are easy to degrade and complex structured and high molecular weight dyes are hard to degrade. The dye decolorization decreased with the increase in initial dye concentration (Liu et al. 2016; Li et al. 2019). In (figure 3), the maximum dye concentration of 400 mg/L showed less dye decolorization when compared to 200 mg/L and 50 mg/L. Similar result trend was reported in the previous studies (Patil et al. 2016; Barathi et al. 2020).
Figure 3: Percentage dye decolorization for different initial dye concentration
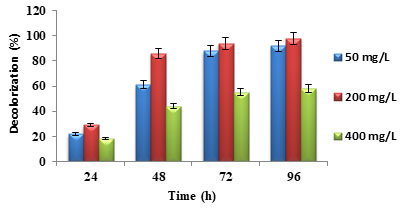
Effect of inoculum size: Inoculum size plays a crucial role in the dye decolorization present in industrial effluents. Larger inoculum size increased dye decolorization while smaller inoculum size showed lesser dye decolorization (Kisand et al. 2001). The same trend was observed in the decolorization of auramine O dye, as shown in figure 4. It was observed that dye decolorization was more for 10% inoculum size when compared to 2% inoculum size. Similar result trend was reported in the previous studies (Roy et al. 2018; Aktar et al. 2019; Shet et al. 2020).
Figure 4: Percentage dye decolorization for different inoculum size
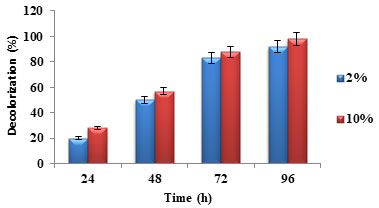
Effect of nitrogen and carbon sources: Nitrogen and carbon supplements are required by the microorganisms for rapid decolorization of dyes (Varjani and Upasani 2019). Organic sources such as yeast extract, peptone and the combination of complex organic sources and carbohydrates have been reported for high and rapid dye decolorization rates by both pure and mixed cultures (Kisand et al. 2001; Nakkeeran et al. 2020).
Figure 5: Percentage dye decolorization for different nitrogen sources
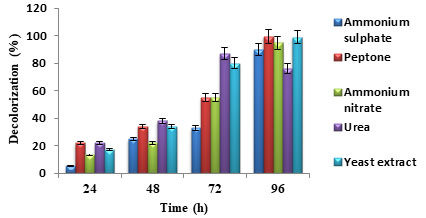
Figure 6: Percentage dye decolorization for different carbon sources
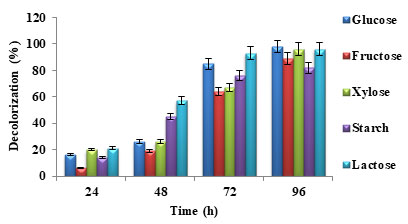
In figure 5, it was observed that nitrogen source peptone favoured maximum dye decolorization efficiency followed by yeast extract. In the case of carbon source, glucose favoured maximum dye decolorization followed by lactose as shown in figure 6. Similar results were reported in the previous studies (Patil et al. 2016; Barathi et al. 2020). For the microbial metabolism of dyes and dyes intermediates, the easily available and effective carbon source is glucose (Khan et al. 2012; Achappa et al. 2021).
CONCLUSION
The findings of the present study deal with degradation of auramine O dye present in textile industry wastewater before disposal to water bodies which otherwise affects the environment. Thus, the degradation of dye present in the industrial effluent is essential and mandatory. The biological treatment method offers the prospective benefits when compared to physical and chemical treatment methods. The potential bacterial isolates were isolated and screened from the effluents of the textile industry.
The gram staining indicated that the culture is a mixed culture that contained both gram-positive and gram-negative organisms. The physical and biochemical tests of the culture confirmed bacillus and staphylococcus species. Optimization studies showed that initial pH, inoculum size, nitrogen source, incubation temperature, carbon source, and initial dye concentration significantly affected the degradation potential of bacteria. Therefore, bacterial strains can be used successfully for the degradation of textile dye basic yellow auramine O.
ACKNOWLEDGEMENTS
This study was supported by the Department of Biotechnology, K. L. E. Technological University, Hubballi, Karnataka, India. Authors thank the department for providing lab facilities to carry out this research work. Moreover, Anil R. Shet and Laxmikant R. Patil have designed the experimental work and executed the experiments. Anil R. Shet, Laxmikant R. Patil, Veeranna S. Hombalimath, and Santosh A. Kadapure have carried out data analysis and interpretation. Anil R. Shet has written the manuscript. Sharanappa Achappa and Shivalingasarj V. Desai have done the grammer check, plagiarism check and proof reading. The manuscript has been approved by all the authors.
Conflict of Interests: Authors declare no conflicts of interests to disclose.
REFERENCES
Achappa, S., Hombalimath, V. S., Kamaraddi, J. R., et al. (2021). Statistical optimization of lipase production from bacillus species by submerged fermentation, Bioscience Biotechnology Research Communications, 14(1), pp. 264-269. http://dx.doi.org/10.21786/bbrc/14.1/37.
Achappa, S., Shet, A, R., Patil, L. R., et al. (2020). Biosynthesis of silver nanoparticles using citrus sinensis peel extract and their application as antibacterial agent, International Journal of Research in Pharmaceutical Sciences, 11(3), pp. 4726-4732. https://doi.org/10.26452/ijrps.v11i3.2762.
Ajaz, M., Shakeel, S. and Rehman, A. (2020). Microbial use for azo dye degradation-a strategy for dye bioremediation, International Microbiology, 23 (2), pp. 149–159.
Aktar, K., Zerin, T. and Banik, A. (2019). Biodegradation of textile dyes by bacteria isolated from textile industry effluent, Stamford Journal of Microbiology, 9(1), pp. 5-8. https://doi.org/10.3329/sjm.v9i1.45649.
Al-Amrani, W.A., Lim, P.E., Seng, C.E., et al. (2014). Factors affecting biodegradation of azo dyes and C.O.D. removal in anoxic-aerobic react operated sequencing batch reactor, Journal Taiwan Institute of Chemical Engineers, 45 (2), pp.609–616.
Barathi, S., Aruljothi, K. N., Karthik, C., et al. (2020). Optimization for enchanced ecofriendly decolorization and detoxification of reactive blue 160 textile dye by Bacillus subtilis, Biotechology Reports, 28: e00522.
Basutkar, M.R. and Shivannavar, C.T. (2019). Decolorization Study of Reactive Red-11 by using Dye Degrading Bacterial Strain Lysinibacillus boronitolerans CMGS-2, International Journal of Current Microbiology and Applied Science, 8 (6), pp.1135–1143.
Bencheqroun, Z., Mrabet, I.E., Kachabi, M., et al. (2019). Removal of basic dyes from aqueous solutions by adsorption onto Moroccan clay (fezcity), Mediterranean Journal of Chemistry, 8 (2), pp.158–167.
Benkhaya, S., M’rabet, S. and El Harfi, A. (2020). Classifications, properties, recent synthesis, and applications of azo dyes, Heliyon, 6 (1), e03271. https://doi.org/10.1016/j.heliyon.2020.e03271.
Bhavikatti, J. S., Bodduchari, S. M., Kamagond, R. S., et al. (2020). Statistical optimization of protease production using a freshwater bacterium Chryseobacterium cucumeris SARJS-2 for multiple industrial applications, 3Biotech, 10:279. doi.org/10.1007/S13205-020-02259-5.
Cao, J., Sanganyado, E., Liu, W., et al. (2019). Decolorization and detoxification of Direct Blue 2B by indigenous bacterial consortium, Journal of Environment Management, 242, pp. 229–237.
Crini, G. and Lichtfouse, E. (2018). Advantages and disadvantages of techniques used for wastewater treatment, Environmental Chemical Letters, 17, pp. 145–155.
Hombalimath, V. S., Udapudi, B. B., Patil, L. R., et al. (2012). Isolation and characterization of lipolytic microorganisms from oil contaminated soil, International Journal of Advances in Engineering, Science and Technology, 2(3), pp. 293-297.
Jamee, R. and Siddique, R. (2019). Biodegradation of synthetic dyes of textile effluent by microorganisms: An environmentally and economically sustainable approach, European Journal of Microbiology and Immunology, 9(4), pp. 114-118. https://doi.org/10.1556/1886.2019.00018.
Khan, R., Bhawana, P. and Fulekar, M.H. (2012). Microbial Degradation and Decolorization of synthetic dyes: a review, Reviews in Environmental Sciences and Biotechnology, 12 (1), pp.75–97.
Kiayi, Z., Lotfabad, T.B., Heidarinasab, A., et al. (2019). Microbial Degradation of azo dye carmoisine in aqueous medium using Saccharomyces cerevisiae ATCC 9763, Journal of Hazardous Materials, 373, pp. 608–619.
Kisand, V., Tuvikene, L. and Noges, T. (2001). Role of phosphorus and nitrogen for bacteria and phytoplankton development in a large shallow lake, Hydrobiology, 457, pp.187–197.
Kulkarni, A., Desai, S. V. and Shet, A. R. (2017). Isolation and characterization of pigment producing actinomycetes from different sources, Research Journal of Pharmaceutical Biological and Chemical Sciences, 8(3S), pp. 101-109.
Li, H., Wang, Y., Wang, Y., et al. (2019). Bacterial Degradation of anthraquinone dyes, Journal of Zhejiang University Science-B, 20 (6), pp.528–540.
Liu, N., Xie, X., Yang, B., et al. (2016). Performance and microbial community structures of hydrolysis acidification process treating azo and anthraquinone dyes in different stages, Environmental Science and Pollution Research, 24 (1), pp. 252–263.
Nakkeeran, E., Varjani, S., Goswami, S., et al. (2020). Chitosan based silver nanocomposite for hexavalent chromium removal from tannery industry effluent using a packed bed reactor, Journal of Environmental Engineering, 146 (6), 04020032. https://doi.org/10. 1061/(ASCE)EE.1943-7870.0001701.
Patil, L. R., Shet, A. R., Lohar, A. G., et al. (2020). Optimization of process parameters for synthesis of silver nanoparticles using leaf extract of tridax procumbent and its biotechnological applications, International Journal of Scientific & Technology Research, 9(6), pp. 1050-1056.
Patil, N. P., Bholay, A. D., Kapadnis, B. P., et al. (2016). Biodegradation of model azo dye methylred and other textile dyes by isolate Bacillus circulans NPP1, Journal of Pure and Applied Microbiolology, 10(4), pp. 2793-2800.
Pratiwi, D., Prasetyo, D.J. and Poeloengasih, C.D. (2019). Adsorption of Methylene Blue dye using Marine algae Ulva lactuca, I.O.P. Conference Series.: Earth and Environmental Science, 251, 012012. https://doi.org/10.1088/1755-1315/251/1/012012.
Roy, D.C., Biswas, S.K., Saha, A.K., et al. (2018). Biodegradation of Crystal Violet dye by bacteria isolated from textile industry effluents, Peer Journal, 6, e5015. https://doi.org/10.7717/preetj.5015.
Shet, A. R., Kouloorkar, S. S., Moolya, D. M., et al. (2013). Production and Characterization of Biodiesel from Cottonseed oil, International Journal of Advances in Engineering, Science and Technology, 2(4), pp. 328-334.
Shet, A. R., More, S., Patil, L. R., et al. (2020). Immobilization of xylanase in PVA-alginate matrix and its characterization, World Journal of Pharmaceutical and Life Sciences, 6(3), pp. 88-91.
Shet, A. R., Patil, L. R., Hombalimath, V. S., et al. (2021). Parametric optimization of oil extraction and lipase catalysed biodiesel production from rice bran, Bioscience Biotechnology Research Communications, 14(1), pp. 340-345. http://dx.doi.org/10.21786/bbrc/14.1/48.
Shet, A. R., Tantri, S. and Bennal, A. (2016). Economical biosynthesis of silver nanoparticles using fruit waste, Journal of Chemical and Pharmaceutical Sciences, 9(4), pp. 2306-2311.
Singh, S., Lo, S.L., Srivastava, V.C., et al. (2016). Comparative study of electrochemical oxidation for dye degradation: parametric optimization and mechanism identification, Journal of Environmental Chemical Engineering, 4 (3), pp. 2911–2921.
Tochhawng, L., Mishra, V.K., Passari, A.K., et al. (2019). Endophytic Fungi: Role in Dye Decolorization, Advances in Endophytic Fungal Research, Fungal Biology, Springer, pp. 1–15.
Varjani, S. and Upasani, V. N. (2019). Influence of abiotic factors, natural attenuation, bioaugmentation and nutrient supplementation on bioremediation of petroleum crude contaminated agricultural soil, Journal of Environmental Management, 1; 245, pp. 358-366. doi: 10.1016/j.jenvman.2019.05.070.
Varjani, S. J. (2017). Microbial degradation of petroleum hydrocarbons, Bioresource Technology, 223, pp. 277-286. doi: 10.1016/j.biortech.2016.10.037.
Varjani, S. J. and Upasani, V. N. (2016). Biodegradation of petroleum hydrocarbons by oleophilic strain of Pseudomonas aeruginosa NCIM 5514, Bioresource Technology, 222, pp.195-201. doi: 10.1016/j.biortech.2016.10.006.


