PG and Research Department of Zoology, Chikkanna Government Arts College, Tirupur-641602, Tamilnadu, India
Article Publishing History
Received: 20/05/2016
Accepted After Revision: 22/06/2016
Pseudomonas spp. has become increasingly recognized as an emerging opportunistic pathogen. A total of 80 isolates of Pseudomonas spp. were isolated from blood cancer patient from three tertiary hospitals in and around Tirupur and Erode (Dts). The pathogenic strains were subjected to antibiotic susceptibility testing by disk diffusion method. Ten different antibiotic discs were used to determine the drug resistance pattern of the isolates. The antibiotic resistant pattern showed that Pseudomonas spp. had high resistant against Imipenem (65%), Co-Trimoxazole (55%), Kanamycin (55%), Amikacin (55%), Erythromycin (55%), Carbenicillin (55%), Piperacillin (55%), Tetracycline (55%), Levofloxacin (50%), and Vancomycin (50%), respectively. The isolates were treated with different metals: Copper, Lead and Zinc with different concentrations (0.2, 0.4, 0.6 and 0.8 mg/ml). Electrophoretic analysis of the Plasmid DNA prepared was carried out on a 0.8% agarose gel electrophoresis. Plasmid profile was carried out for 22 MDR isolates. Four strains (PA 41, PA 43 and PA 44) were showed two fragments (10,000 bp). Whereas the isolate (PA 47) showed single plasmid fragment (6,000 bp).
Antibiotics, Blood Cancer, Heavy Metals, Pseudomonas Spp.
Rajasekar S, Mohankumar A. Antibiotic Susceptibility and Plasmid Profile of Heavy Metal Resistant Pseudomonas Species. Biosc.Biotech.Res.Comm. 2016;9(2).
Rajasekar S, Mohankumar A. Antibiotic Susceptibility and Plasmid Profile of Heavy Metal Resistant Pseudomonas Species. Biosc.Biotech.Res.Comm. 2016;9(2). Available from: https://bit.ly/2MLcEBl
Introduction
Cancer patients with numerous types of malignancies often become infected with Pseudomonas spp. A total of 25% of all gram negative bacillary infection in cancer patients are due to Pseudomonas spp. the majority occurring in patients with neutropenia (Rodriguex and Bodey, 1979). Pseudomonas spp. is an epitome of opportunistic nosocomial pathogen, which caused a wide spectrum of infection and leads to substantial morbidity in immune compromised patients despite therapy the mortality due to nosocomial Pseudomonas spp. is approximately 70%. Most of the genetic determinants that confer resistance to antibiotics are located on plasmids. These extra-chromosomal DNA sequences are often transferable to other bacteria in the environment and can be responsible for the emergence of multiple resistances to antibiotics (Schelz et al., 2006). In recent years heavy metal tolerance isolates have showed maximum microbial tolerance to Cr and minimum tolerance to Pb and Cu (Singh and Lal, 2015).
The heavy metal and radioactive pollution from nuclear power plants, mining industries, electroplating industries and agricultural runoffs is a major cause of concern to public health, animals and ecosystem (Rehman et al., 2007). Lead is hazardous waste and highly toxic to human plants animals (Low et al., 2000). One of the major mechanisms by which heavy metals including zinc cause toxicity is through the generation of free radicals and oxidative (Krulwich et al., 2005). Resistance to antibiotics can be conferred by chromosomal or mobile genetic elements (e.g. plasmids) and achieved using four main strategies: reduction of membrane permeability to antibiotics, drug inactivation, and rapid efflux of the antibiotic and mutation of the cellular target (Giller and Witter, 1988). Some heavy metal resistance determinants move from plasmid to chromosome.
The Pseudomonas spp. ever growing multi drug resistance has been widely reported (Agarwal et al., 2005). This makes plasmid encoding heavy metal resistance an important aspect of environmental research. Therefore, this present investigation has made an attempt to point out the antibiotic resistance of carcinogenic organism which is isolated from different blood cancer patients from different tertiary hospitals in and around Tirupur and Erode district, Tamilnadu and the multidrug resistant organism link with heavy metal resistant plasmid of cancer causing Pseudomonas spp.
Materials And Methods
Bacterial Strains And Culture Conditions
A total of 80 Pseudomonas spp. isolates, isolated from blood cancer patients in and around Tirupur District. Isolation was made by adding 5ml blood obtained from peripheral veins of the patients to brain heart infusion (BHI) broth. It was incubated at 37°C and regular subcultures were done. Identification of the Pseudomonas spp. isolates was done by morphological and standard biochemical tests according to the manual of clinical microbiology (Cheesbrough, 1984). Identified Pseudomonas spp. strains were stored as glycerol stocks at -20°C until use.
Antibiotic susceptibility Test 10 antibiotics were used to determine antibiotic resistance of carcinogenic stains. These antibiotic discs were Imipenem (10mcg), Amikacin (10mcg,) Co-Trimoxazole (30mcg), Kanamycin (10mcg), Piperacillin (30mcg), Tetracycline (30mcg), Vancomycin (30mcg), Carbenicillin (30mcg), Erythromycin (15mcg) and Levofloxacin (5mcg). The susceptibility tests for each isolates were performed using Kirby Bauer disk diffusion method. The disk were placed on the solidified agar surface. The plates were incubated aerobically for 24 h at 37°C. The resistance were determined according to the zone of inhibition by the isolates.
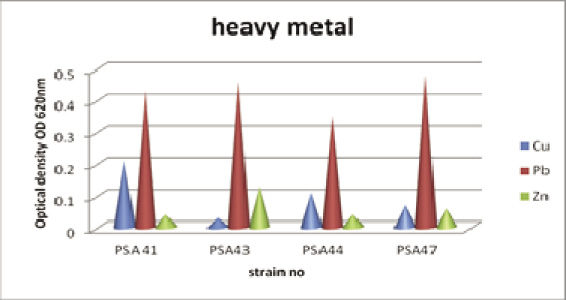 |
Figure 1 |
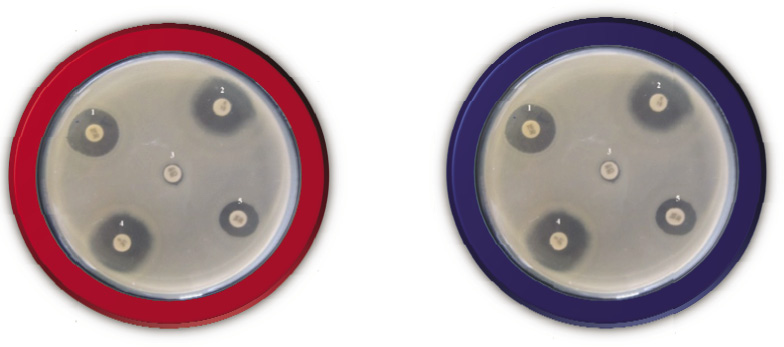 |
Figure 2 |
Growth Studies
Growth studies of clinical bacterial isolates were studied in 250 ml flasks containing 50 ml Luria Broth medium supplemented with 0.1mM concentration of different metals such as Copper, lead and Zinc. 0.5 ml of overnight culture was added to conical flask containing culture medium. It was kept in Rotary shaker at 150 rpm, 24 h. The growth of carcinogenic pathogen was checked by absorbance at 620 nm using spectrophotometer (Selvam et al., 2009).
Plasmid Dna Isolation
Plasmid DNA was isolated based on alkaline lysis method described by (Kado et al., 1981). Purified DNA preparations were analyzed on 0.8% agarose gels stained with ethidium bromide and visualized using (Maniatis et al., 1982).
Results And Discussion
Totally 80 multi drug resistant isolates were identified as Pseudomonas spp., according to Bergey’s Manual of systematic bacteriology. The isolated carcinogenic isolates were tested to determine the antibiotic resistant isolates by Disc diffusion assay. The antibiotic discs of Imipenem (10mcg) Amikacin (10mcg), CO-Trimoxazole (30mcg) Kanamycin (10mcg), Piperacillin (30mcg), Tetracycline (30mcg), Vancomycin (30mcg), Carbenicillin (30mcg), Erythromycin (15mcg), and Levofloxacin (5mcg). Isolate were analyzed for to determine the antibiotic susceptibility pattern along with the tendency of current resistance against widely used drugs among 80 isolates.
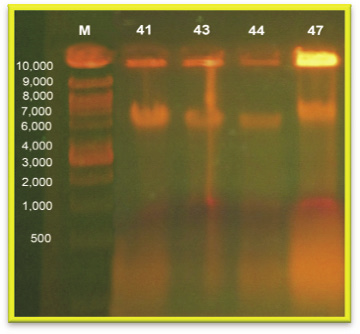 |
Figure 3 |
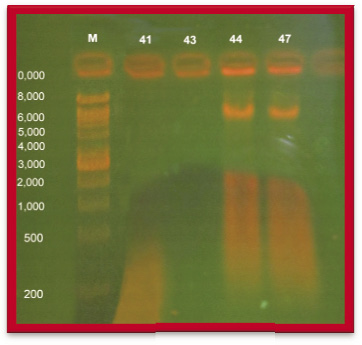 |
Figure 4 |
Among 80 isolates 30 different antibiogram were found and showed resistance against imipenem (65%), CO-Trimoxazole (55%), Kanamycin (55%), Amikacin (55%), Vancomycin (50%), Carbenicillin (55%), Levofloxacin (50%), Erythromycin (55%), Piperacillin (55%), and Tetracycline (55%).
Isolates which are showed (50%), resistant against tested antibiotics were used for plasmid DNA isolation by alkaline lysis method. Two fragment were obtained in strain No. PA 41, PA 43, PA 44 and PA 47 with molecular size of 10,000 bp and 7000 bp respectively.
The highest level of resistance carcinogenic isolates were treated with metal curing agents such as Lead, Zinc and Copper with different concentration (0.2%, 0.4%, 0.6% and 0.8% mg∕ml). Cured colonies of Pseudomonas spp. were taken for the isolation of plasmid. According to four multidrug plasmid isolates, two isolates PA 41, PA 43 possessed no plasmid it denotes that the cancer causing Pseudomonas spp. was most sensitivity to curing agents such as Zinc, Copper and Lead and other two heavy metals resistant PA 44 and PA 47 harbour two fragments of molecular size between 10, 000 bp and 6, 000 bp respectively.
Akingbade et al. (2012) have showed Pseudomonas spp. had high resistant to Amoxicillin (92.7%), Ampicillin (90%), Cloxacillin (88.2%), Cotrimoxazole (77.3%), Erythromycin (72.7%), Tetracycline (70.9%), Streptomycin (65.5%), and Ofloxacin 60% and had low resistant to Ceftazidime (20%), Gentamycin (26.4%), Levoxin (30.9%), Ceftriaxone (34.5%) and Ciprofloxacin (35.5%). Further they carried out the plasmid profile on 22 selected multi drug resistant (MDR) isolates that were resistant to three or more classes of antibiotics. Eight (36.4%) strains were found to possess plasmid born. Six of the strains had single plasmid band while two strains possessed two bands with sizes ranging from 662 to 830bp. The sizes of the plasmids among Pseudomonas spp. isolates ranged from 662bp to 830bp. All the strains that had plasmids were resistant to Amoxicillin, Ampicillin, Cloxacillin, Cotrimoxazole, Erythromycin and Tetracycline. In this present investigation also followed the similar method for antibiotic susceptibility test. But dissimilar results were found that the antibiotic resistant pattern showed that Pseudomonas spp. had low resistant to Co-Trimoxazole (55%), Tetracycline (55%) and Erythromycin 55%. Electrophoretic analysis of plasmid DNA was carried out 0.8% agarose gel analysis. Plasmid profile was carried out on 4 selected multi drug resistant (MDR) isolates that were resistant to four group of antibiotics. Four strains were found to possess plasmid born. According to all four strains harbour two plasmid bands with molecular size between 10,000 bp – 7000 bp respectively.
The antibiotic resistance of clinical pathogen is emerging in hospital settings to adequately diagnosis pseudomonal infections and prescribe the antibiotic treatment most effective in preventing the increase in multidrug resistant organisms (Brown and Anicetus Izundu, 2004). Organisms were cultured from wound swabs (56%), high vaginal swabs (10.5%) and ear swabs (42.5%). Overall, the highest percentage rates of resistance were found for cofactor (100% of all isolates), Nalidixic acid (82.4%), Kanamycin (76.5%), and Trimethoprim/Sulfamethoxazole (56.9%). Resistance rates were (25.5%) or lower for Tobramycin, Gentamicin and Polymyxin B, Cefotaxime, Ciprofloxacin and Norfloxacin, Piperacillin, Carbapenems and Amikacin. Forty-one isolates showed intermediate sensitivity to most of the anti pseudomonal antibiotics, and the remaining 10 isolates were resistant to eight or more antibiotics. The multi resistant isolates showed resistant to Tetracycline, Nalidixic acid and Trimethoprim/Sulfamethoxazole, and highly (80%–90%) resistant to Kanamycin, Ciprofloxacin and Norfloxacin. In this research, dissimilar results were found in all the Pseudomonas spp. microbes in blood cancer origin were high resistant to Piperacillin (55%), Amikacin (55%) and low resistant percentage rates were found in Kanamycin (55%), and Tetracycline (55%).
Abdel-Salam et al., (2007) examined forty nine Pseudomonas aeruginosa against ten heavy metals. They were showed resistant to seven to nine heavy metals and four to five antibiotics. The ten strains contain two small plasmid fragments, but have up to five large plasmids. Plasmid curing indicated that resistant to each of Cadmium, Cobalt, Silver, Chromium, Mercury, Nickel or Zinc is plasmid borne. It also showed that the resistance of both iron and lead are carried by the bacterial chromosome. Results also revealed that chromate resistance genes were located on a large plasmid in Pseudomonas 24 strain. The resistance of all tested antibiotic was located in plasmids. Similar results was obtained in this present study the isolates of Pseudomonas spp. isolates from blood cancer patient and were examined for resistant against three heavy metals and ten antibiotics. Plasmid profile was carried out on 4 selected multi drug resistant (MDR) isolates that were resistant to four group of antibiotics. Four strains were found to possess plasmid bands. Total four MDR strains harbour two bands. Plasmid curing indicated that resistant to Zinc is plasmid borne. It also showed that the resistance of lead also carried by the bacterial chromosome.
Mathiyazhagan and Natarajan, (2012) have focused on the isolation of metal tolerant and antibiotic sensitive bacteria: A. ferrooxidans from bauxite and P. aeruginosa by using selective medium. These two organisms showed maximum metal resistant potentiality for the selected heavy metals (Mn, Zn, Fe, Cr, Cu and Hg). In this present investigation the cancer causing Pseudomonas spp. isolates were isolated from blood cancer patient using selective medium. The isolates were treated with different metals Copper, Lead, and Zinc with different concentrations (0.2, 0.4, 0.6 and 0.8 μg/ml). The antibiotic susceptibility of (50%) resistant strains of were used for metal tolerant.
Conclusion
The present study concluded that, there is tremendous multiple drug resistance found to different class of antibacterial drugs. There is a quite alarming signal to note that all isolates of antibacterial Pseudomonas spp. were resistance to more antibiotics. All the isolates showed multiple drug resistance characteristics and this organism link with plasmid encoding heavy metal resistant features.
Acknowledgements
The authors wish to thank Dr. K. Shanmugasundharam, Principal, Chikkanna Government Arts College, and Tirupur for providing facilities to conduct the research.
References
Abdel-Salam M.S., A. Nivien Abosereh, A.Z. Abd El-Salam, Ibrahim S.A. and Saeb, A.T.M. (2007). Resistance Plasmids of Indigenous Pseudomonas in Egypt. Journal of Applied Sciences Research., Vol. 3, 873-878.
Agarwal G., A. Kapil, S.K. Kabra, Das B.K. and Dwived D. (2005). Characterization of Pseudomonas isolated chronically infected children cystic fibrosis in Indian. BMC Microbiology, Vol. 5, 43.
Akingbade O.A., S.A Balogun, D.A Ojo, R.O. Afolabi, B.O. Motayo, P.O. kerentugba, I.O. and Okonko (2012). Plasmid Profile Analysis of Multidrug Resistant Pseudomonas aeruginosa isolated from Wound Infection in South West, Nigeria. World applied science Journal., Vol 20, Issue 6, 766-775.
Brown P.D., and Izundu A. (2004). Antibiotic resistance in Clinical isolate of Pseudomonas aeruginosa in Jamaica Revmpanam salud publica, Vol. 16, Issue 2, 125-30
Edward Raja C., Anbazhagan K and Selvam G.S, (2006). Isolation and characterization of a metal resistant Pseudomonas aeruginosa strain. World Journal of Microbiology and Biotechnology. Vol. 22, 577- 586.
Giller KG., Witter E and McGrath S.P. (1998). Soil Biol Bioch, Vol 30, 1389–1414.
Kado C.I., and Liu S.T. (1981). Rapid procedure for detection and isolation of large and small plasmids. J. Bacteriol., Vol.145, 1365-1373.
Krulwich TA., and Lewinson O. (2005). Do physiological roles foster persistence of drug/multidrug- efflux transporters? A case study. Nat. Rev. Microbiol. Vol. 36, 566-572.
Low K.S., Lee C.K. and Liew S.C. (2000). Sorption of Cadmium and Lead form aqueous solution by spent grain. Proc. Biochemist., Vol.36, Issue 1-2, 59-64.
Maniatis T. E. F., Fritsch and Sambrook J. (1982). “Molecular Cloning, A Laboratory Manual”, Cold Spring Harbor, New York.
Narayanan Mathiyazhagan and Devarajan Natarajan, (2012). Metal and Antibiotic Tolerance Potentiality of Acidithio bacillus Spp. and Pseudomonas spp. from waste Dumps of bauxite and magnesite Mines. Archives of Applied Science Research, vol 4, Issue 1, 616-622.
Rehman A and Shakoori FR. (2007). Multiple metal resistance and uptake by a ciliate, Stylonychia mytilus, isolated from industrial effluents and its possible use in wastewater treatment. Bull. Environ. Contam. Toxicol., Vol 79, Issue4, 410-414.
Rodriguez V and Bodey G. P. (1979). Epidemiology, clinical manifestations and treatment in cancer patients in R. G. Dogget (ed.), Pseudomonas aeruginosa– clinical manifestations of infection and current therapy. Academic Press, Inc., New York, 367–407.
Schelz Z., Molnar J and Hohmann. J (2006). Antimicrobial and anti plasmid, activities of essential oils. Fitoterapia, Vol 77, 279-8.
Yogendra Singh and Nand Lal (2015). Investigation on the heavy metal resistant bacterial isolates in vitro from industrial effluents. World Journal of Pharmacy and Pharmaceutical Science. Vol 4, Issue 02, 345 -350.


