1Bioinformatics and Supercomputer Laboratory, Department of Biosciences (UGC-SAP-DRS-II & DST-FIST-I),
Veer Narmad South Gujarat University, Surat-395007, India.
2 Shri Alpesh N. Patel Post Graduate Institute of Science and Research,
Department of Biochemistry, Anand-388001, India.
3Smt GBP & Smt PSP Sciences (M.Sc) College, Palanpur-385001, India.
Corresponding author email: hnguraj@gmail.com
Article Publishing History
Received: 03/10/2022
Accepted After Revision: 20/12/2022
Panchagavya is the blend of five ingredients obtained from cows and used in traditional Hindu rituals. Panchagavya preparation is a microbial mediated process that possibly involves microbial succession. The present study was conducted to decode the microbial community that exists in the preparation of three hours of old Panchagavya. DNA was isolated from Panchagavya using HiPurA™ Stool DNA Purification Kit followed by quality checking by Agarose electrophoresis and Qubit fluorometer. The V3 and V4 region of 16s rRNA based amplicon sequencing was performed using the Illumine MiSeq platform. Taxonomic profile encoded by using One Codex, kraken and MG-RAST. Functional traits detected through the abundance of specific genes using Tax4Fun. Taxonomic result suggests the total 2000 species were identified.
The most abundant was Streptomyces griseocarneus (2.65%) followed by Clostridiales bacterium (2.26%), Bacteroidales bacterium (1.38%), and Verrucomicrobia bacterium (1.13%). Community based analysis revealed the microbial diversity and presence of anaerobic, unclassified, and uncultivable microbes in metagenomes, which may be associated with the pharmacological properties of Panchagavya. Functional analysis predicts around 351 metabolic pathways for metabolism of carbohydrates, synthesis of secondary metabolites and degradation of xenobiotic compounds. The detection of various secondary metabolites genes associated with pharmacological molecules correlated with its traditional clinical applications. The present study revealed the advantages of cultivation approach for exploring untapped and unique bacterial diversity, and also utilities for various biotechnological and environmental applications.
Amplicon Sequencing, Community Metagenomics, Cow, Functional Metagenomics, Panchagavya
Vaghamshi N, Gandhi H, Beladiya U, Mangrola A, Dudhagara P, Patel D.R, Chaudhari R. 16s rRNA Amplicon Sequencing Approach for Community and Predictive Functional Diversity of Therapeutically Valuable Formulation of Cow-derivatives. Biosc.Biotech.Res.Comm. 2022;15(4).
Vaghamshi N, Gandhi H, Beladiya U, Mangrola A, Dudhagara P, Patel D.R, Chaudhari R. 16s rRNA Amplicon Sequencing Approach for Community and Predictive Functional Diversity of Therapeutically Valuable Formulation of Cow-derivatives. Biosc.Biotech.Res.Comm. 2022;15(4). Available from: <a href=”https://bit.ly/3C3qB8l“>https://bit.ly/3C3qB8l</a>
Copyright © This is an Open Access Article distributed under the Terms of the Creative Commons Attribution License (CC-BY). https://creativecommons.org/licenses/by/4.0/, which permits unrestricted use distribution and reproduction in any medium, provided the original author and sources are credited.
INTRODUCTION
Panchagavya is an organic product derived from five products of the cow. The three are direct constituents i.e. dung, urine and milk and the two derived products are curd and ghee. According to the old Ayurveda literature, the five individual constituents of Panchagavya possess medicinal properties and can be used singly or in combination with some other herbs. This kind of treatment is called Panchgavya therapy or cowpathy (Muthukapalli et al., 2022).
The potential applications of Panchagavya are as antimicrobials, immune boosters, antidiabetics, anticancer, anticonvulsant, aphrodisiac, blood purifiers, and anti-HIV agents. Panchagavya is also used in Ayurveda for the treatment of several disorders such as hyperlipidemia, leucoderma, arthritis, acidity, renal disorders, asthma, dietary disorders, gastrointestinal disorders and asthma as an antistressor. It also acts as a general tonic or immunomodulator to enhance the immunity (Dhama et al., 2022, Athavale et al., 2012 & Kuldeep et al., 2013). Panchagavya is essential in organic agriculture for disease control, plant growth, flowering, germination, and canopy development. It also plays a significant part in compost decomposition, improves soil nutrient status by promoting better mineralization, and aids in the creation of beneficial microbes in the root rhizosphere, which can helpful in the supply of beneficial macro and micro nutrients to plants (Kumar et al., 2022).
Panchagavya contains several macronutrients such as nitrogen, phosphorus, potassium and micronutrients like Zn, Fe, Cu, and Mn which are required for the normal growth and development of plants. In the Indian subcontinent, the application of the Panchagavya is well-known and used in an agricultural sector to protect the crops and growth promotion of the crops (Gugalia et al., 2021, Kumaravelu et al., 2009, Dhama et al., 2005, Chauha, R.S., 2002, Krupanidhi et al., 2008, Matthews and Jenks, 2013, Kumar et al., 2022).
By culture dependent study few bacterial species isolated from the Panchagavya were Lactobacillus, Saccharomyces, Streptomyces, and Rhodopseudomonas (Leo et al., 2013), Azospirillum, Azotobacter, phosphobacteria (Dhama et al., 2013), Pseudomonas, Azotobacter, Actinomycetes, Rhizobium (Ram et al., 2013), Acetobacter, Bacillus, Micrococcus, Leuconostoc, Enterococcus, Microbacterium, Pseudoxanthomonas, Corynebacterium, Escherichia, Paenibacillus, Shigella, Rhodobacter, Lactococcus, etc. (Anandham et al., 2015)0.
Nowadays, the Prophylactic potential of a Panchagavya formulation was tested against certain pathogenic bacteria (Patel et al., 2018). So, based on such diverse properties and the presence of different bacterial species; there a need to decode the microbiome of the Panchagavya formulation. To the best of our knowledge; at present, there is not a single study of the metagenomics analysis of Panchagavya. This is the first microbial profiling of the Panchagavya by culture independent methods using a metagenomics approach.
MATERIAL AND METHODS
Panchagavya was prepared at Madhuvan Dairy Farm, Haldarva, Bharuch, India. Fresh cow dung, urine, milk, curd, and ghee were mixed in equal proportion thoroughly in a sterile glass beaker. This mixture was allowed to stand for three hours and subjected to filtration through a muslin cloth aseptically.
DNA was isolated from the filtrate using HiPurA™ Stool DNA Purification Kit (MB544) according to the manufacturer’s instruction. DNA concentration was measured using the Qubit Fluorometer. The sequencing library generated from V3 and V4 amplicons from the sample were sequenced using an Illumina MiSeq sequencing platform. Diversity and abundance were analysed using available standard bioinformatics software. The taxonomic assignment of unassembled metagenomic sequences was performed using FASTQC followed by Pear, One Codex (Minot et al., 2015) and predictive functional metagenomics performed by SILVAngs, MicrobiomeAnalyst (Dhariwal et al., 2017), Tax4fun (Aßhauer et al., 2015), KO and KEGG mapper (Kanehisa & Sato 2020).
RESULTS
The present study is based on the 16s rRNA amplicon NGS metagenomic analysis of Panchagavya. A total number of 27301 bacterial 16S V3-V4 high- quality sequences with an average read length of 71 to 490, were obtained. Profiling the taxonomic composition of the community also can be accomplished by the analysis of the distribution of k-mers (e.g., using Kraken or One Codex). Metagenomic analysis using One Codex was performed by uploading the 2D FASTQ data to the One Codex platform at https://app.onecodex.com. This cloud-based k-mer method was selected, because it is reportedly more accurate than either the MG-RAST or the Kraken tools and it provides for community access to the data and analytical results. Sequence clustering resulted in the identification of 2000 different bacterial species, 87 diverse microbial phyla and 510 families together with an unclassified category were depicted in the metagenome (Figure 1).
Figure 1: Krona based chart at family level. (Core of chart indicated phyla and peripheral
edge indicates the family’s abundance in percentage)
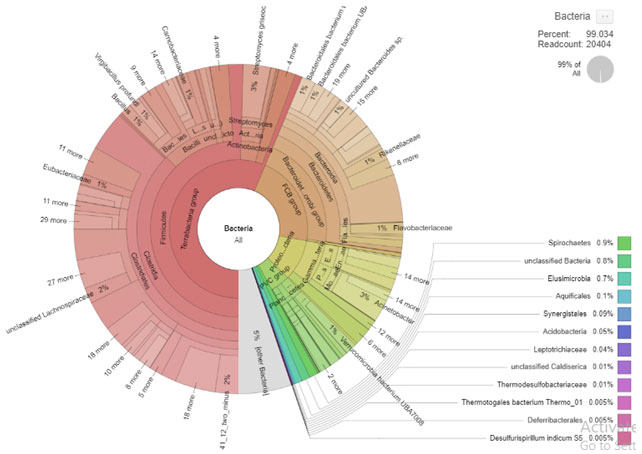
Figure 2: Community at Phyla level based on the percentage of reads in metagenome sample
(Pie chart includes the most 15 dominant phyla presented in clockwise direction)
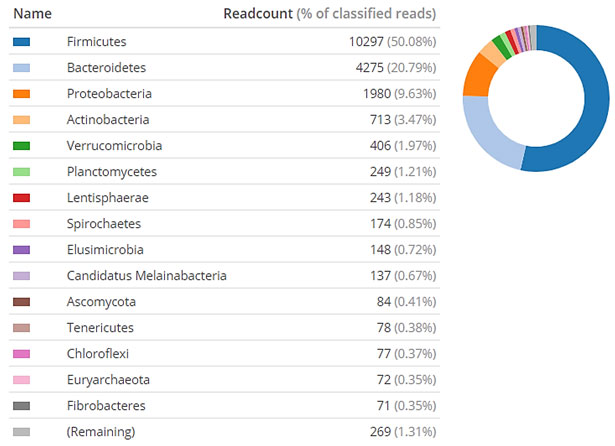
Firmicutes (50.08%) and Bacteroidetes (20.79%) were abundant along with Proteobacteria (9.63%), Actinobacteria (3.47%), Verrucomicrobia (1.97%) and Planctomycetes (1.21%) (Figure 2).
Figure 3: Microbial communities at species level based on the percentage of reads in metagenome
sample (Pie chart includes the most 15 dominant species presented in clockwise direction)
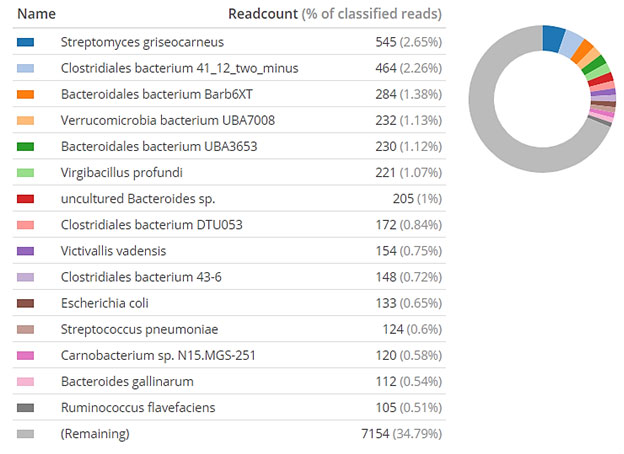
Total of 516 families was detected (figure 1) along with following Ruminococcaceae (8.46%), Lachnospiraceae (7.34%), Bacteroidaceae (3.57%), Enterobacteriaceae (3.27%) and Moraxellaceae (3.12%) were major dominant (Figure 3).
Based on the microbiome analyser, community analysis data was subjected to the functional analysis to decode the functional potential of the microbiome. In the present study pathways modules for the metabolism of cofactor and vitamins, biosynthesis of secondary metabolites, xenobiotic degradation, drug resistance, lipid metabolism, and energy metabolism were detected which correlates its pharmaceutical and agricultural application (Figure 4). Furthermore, we have analysed the 10 categories of Reconstruction Pathway (Table 1).
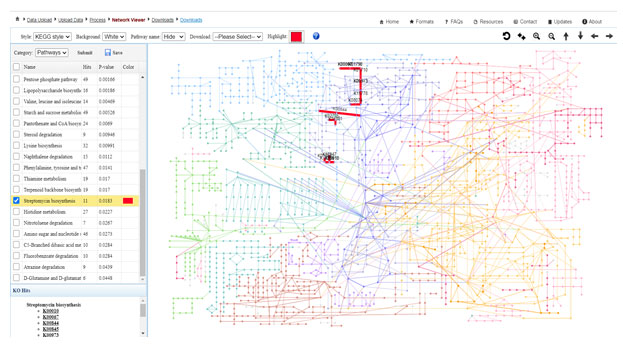
The highest number of the pathway was found in Environmental information processing. To confirm the pharmacological and medicinal attributes of the Panchagavya, we have further analysed the Biosynthesis of other secondary metabolites pathways and were find out the 12 important biosynthetic pathways which give the antimicrobial substances. Furthermore the biosynthesis of Streptomycin was highlighted in the global metabolic network (Figure 5).
Figure 4: Secondary metabolites pathways analysis using KEGG Mapper
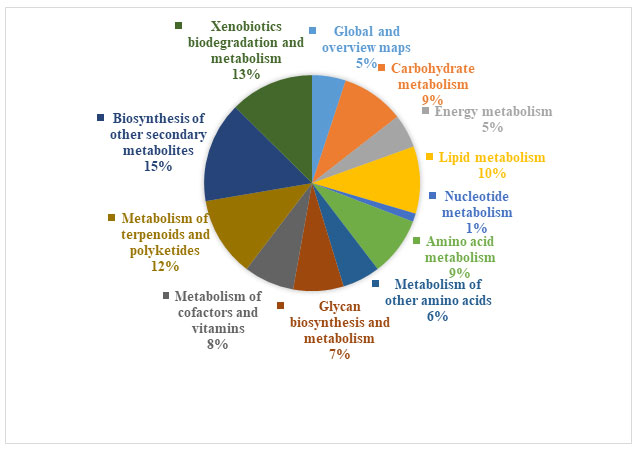
Table 1. List of reconstruction pathway found in the metagenome sample (number of pathways indicates
total different pathways operated in microbiota of sample under each categories)
| Sr. No. | Reconstruction Pathway | No. of Pathway |
| 1 | Carbohydrate metabolism | 41 |
| 2 | Energy metabolism | 62 |
| 3 | Lipid metabolism | 32 |
| 4 | Nucleotide metabolism | 08 |
| 5 | Amino acid metabolism | 53 |
| 6 | Glycan metabolism | 24 |
| 7 | Metabolism of cofactors and vitamins | 31 |
| 8 | Biosynthesis of terpenoids and polyketides | 45 |
| 9 | Xenobiotics biodegradation | 21 |
| 10 | Biosynthesis of other secondary metabolites | 28 |
Figure 5: A depiction showing functional enrichment analysis and visualization within the
global metabolic network. (Highlighted red lines shows the Streptomycin biosynthesis)
DISCUSSION
Panchagavya plays major role in crop production, especially in organic agriculture, maintaining genetic biodiversity, helps as a growth promoter, root growth enhancer, increasing water holding capacity, increase photosynthetic activity in plants, human diseases and improving the body’s immunity, the metabolic profiles (hormones, proteins, nutrients, etc.) and microbial profiles have not been explored completely (Somasundaram et al., 2007; Kumar et al., 2022).
Moreover, the profile and dynamics of the entire microbial community associated with the Panchagavya are unavailable due to limitations in culturing several genera/species using conventional microbiological techniques. Not many efforts have been made to explore the complete biology involved in agriculture, indicating its potential for increasing crop output by the Panchagavya. Only by combining different omics approaches can the fundamental aspects of a complex system be understood (Segate et al., 2013).
Therefore, we have used different omics approaches to decode the microbial community (metagenomics) and metabolite profile (metabolomics) in the Panchagavya. These approaches can reveal the microbial composition and their abundance, the functional annotation of genes and important protein compounds, hormones, and so forth, in the Panchagavya formulation. Several bacteria that used as starter cultures for dairy products were found to be predominant in Panchagavya. Among them Lactococcus termiticola, Lactococcus fujiensis JCM 16395, and Lactococcus fujiensis, is widely used as starter cultures for various cheeses (Lahtinen et al., 2011). Our findings are in contrast to a recent report that the use of Bacillus cereus, Bacillus subtilis, Lactobacillus camellia, Lactobacillus ozone, and Paenibacillus as a probiotic dietary supplements is expanding rapidly with increasing number of studies demonstrating immune stimulation, antimicrobial activities and competitive exclusion (Kamilya et al., 2022; Cutting, 2011; Sorokulova et al., 2008).
CONCLUSION
Using metagenomic sequencing, the Panchagavya’s related microbiota can be profiled. An extensive range of bacterial taxa from the domain bacteria were found in the Panchagavya, according to the research. In Panchagavya metagenomes, the Firmicutes, Bacteroidetes, and Proteobacteria phyla are widely present. Its health benefits are suggested by the presence of a few probiotic species and antibiotic-producing species. The research sheds light on the existence of several bacterial species, whose immune-boosting characteristics and therapeutic potential may be the basis for the Panchagavya’s health-tonic properties. According to pathway research, the secondary metabolites that the bacteria create may be the cause of the Panchagavya’s antibacterial abilities. The Panchagavya is a low-cost instrument for enhancing gut immunity and has a positive impact on health because the enormous microbial variety and the detection of rare species and uncultivable species play a crucial role in maintaining an intestinal barrier and metabolising nutrients.
Data availability statement: Metagenome sequences data are deposited in EBI-Metagenomics under the accession number PRJEB31987
ACKNOWLEDGMENTS
We are extremely grateful to Madhuvan Dairy Farm, Haldarva, Bharuch, India for providing the samples for this study and for sharing knowledge on panchagavya.
Conflict of interest: The authors declare that they have no conflict of interest.
REFERENCES
Anandham, R., Premalatha, N., Jee, H.J., Weon, H.Y., Kwon, S.W., Krishnamoorthy, R., Gandhi, P.I., Kim, Y.K. and Gopal, N.O., (2015). Cultivable bacterial diversity and early plant growth promotion by the traditional organic formulations prepared using organic waste materials. International Journal of Recycling of Organic Waste in Agriculture, 4(4), pp.279-289.
Athavale, A., Jirankalgikar, N., Nariya, P. and Des, S., (2012). Evaluation of in-vitro antioxidant activity of panchagavya: a traditional ayurvedic preparation. Int J Pharm Sci Res, 3, pp.2543-2549.
Chauhan, R.S., (2002). April. Medical importance of Panchgavya (Cow therapy). In National Symposium on Historical Overview on Veterinary Sciences and Animal Husbandry in Ancient India, Vedic and Ashokan Period). IVRI, Izatnagar.
Cutting, S.M., (2011). Bacillus probiotics. Food microbiology, 28(2), pp.214-220.
Dhama, K., Chakraborty, S., Wani, M.Y., Verma, A.K., Deb, R., Tiwari, R. and Kapoor, S., (2013). Novel and emerging therapies safeguarding health of humans and their companion animals: a review. Pakistan Journal of Biological Sciences: PJBS, 16(3), pp.101-111.
Dhama, K., Khurana, S.K., Karthik, K., Tiwari, R., Malik, Y.P.S. and Chauhan, R.S., (2014). Panchgavya: Immune-enhancing and therapeutic perspectives. J Immunolol Immunopathol, 16, pp.1-11.
Dhama, K., Rathore, R., Chauhan, R.S. and Tomar, S., (2005). Panchgavya (Cowpathy): an overview. International Journal of Cow Science, 1(1), pp.1-15.
Gugalia, G., (2021). A Sustainable Agriculture: Organic Farming: A Review. Quarterly Research Journal of Plant & Animal Sciences/Bhartiya Krishi Anusandhan Patrika, 36(3).
Kamilya, D. and Devi, W.M., (2022). Bacillus Probiotics and Bioremediation: An Aquaculture Perspective. In Bacilli in Agrobiotechnology (pp. 335-347). Springer, Cham.
Krupanidhi, K., Kekuda, T.R.P., Bhramarambha, B.K.M., Shrungashree, R.M., Suchitra, S.V. and Kavya, R., (2008). Comparative studies on in vitro Nematicidal (Anthelmintic) activity of Cow urine and Cow urine. Biotechnology: An Indian Journal, 2(1), p.23.
Kuldeep, D., Sandip, C. and Ruchi, T., (2013). Panchgavya therapy (Cowpathy) in safeguarding health of animals and humans-a review. Research Opinions in Animal and Veterinary Sciences, 3(6), pp.170-178.
Kumaravelu, G. and Kadamban, D., (2009). Panchagavya and its effect on the growth of the greengram cultivar K-851. International Journal of Plant Sciences (Muzaffarnagar), 4(2), pp.409-414.
Lahtinen, S., Ouwehand, A.C., Salminen, S. and von Wright, A. eds., (2011). Lactic acid bacteria: microbiological and functional aspects. CRC Press.
Leo Daniel Amalraj, E., Praveen Kumar, G., Mir Hassan Ahmed, S.K., Abdul, R. and Kishore, N., (2013). Microbiological analysis of panchagavya, vermicompost, and FYM and their effect on plant growth promotion of pigeon pea (Cajanus cajan L.) in India. Organic Agriculture, 3(1), pp.23-29.
Matthews, D.M. and Jenks, S.M., (2013). Ingestion of Mycobacterium vaccae decreases anxiety-related behavior and improves learning in mice. Behavioural processes, 96, pp.27-35.
Muthukapalli Krishnareddy, P., Hirehally Basavarajegowda, M., Perumal Buela, P., Devanna, P., Makali Eregowda, P., Sarangi, A.N., Kodihalli Govindaraju, M., Middha, S.K. and Banakar, S.N., (2022), Decoding the microbiome and metabolome of the Panchagavya—An indigenous fermented bio-formulation. iMeta, p.e63.
Patel, P., Joshi, C., Funde, S., Palep, H. and Kothari, V., (2018). Prophylactic potential of a Panchgavya formulation against certain pathogenic bacteria. F1000Research, 7.
Minot, S. S., Krumm, N., & Greenfield, N. B. (2015), One codex: a sensitive and accurate data platform for genomic microbial identification. BioRxiv, 027607.
Dhariwal, A., Chong, J., Habib, S., King, I. L., Agellon, L. B., & Xia, J. (2017), MicrobiomeAnalyst: a web-based tool for comprehensive statistical, visual and meta-analysis of microbiome data. Nucleic acids research, 45(W1), W180-W188.
Aßhauer, K. P., Wemheuer, B., Daniel, R., & Meinicke, P. (2015), Tax4Fun: predicting functional profiles from metagenomic 16S rRNA data. Bioinformatics, 31(17), 2882-2884.
Kanehisa, M., & Sato, Y. (2020), KEGG Mapper for inferring cellular functions from protein sequences. Protein science, 29(1), 28-35.
Ram, R.A., Singha, A. and Vaish, S., (2018), Microbial characterization of on-farm produced bio-enhancers used in organic farming. ICAR.
Segata, N., Boernigen, D., Tickle, T.L., Morgan, X.C., Garrett, W.S. and Huttenhower, C., (2013). Computational meta’omics for microbial community studies. Molecular systems biology, 9(1), p.666.
Somasundaram, E., Amanullah, M.M., Vaiyapuri, K., Thirukkumaran, K. and Sathyamoorthi, K., (2007). Influence of organic sources of nutrients on the yield and economics of crops under maize based cropping system. Journal of Applied Sciences Research, 1774-1777.
Sorokulova, I.B., Pinchuk, I.V., Denayrolles, M., Osipova, I.G., Huang, J.M., Cutting, S.M. and Urdaci, M.C., (2008). The safety of two Bacillus probiotic strains for human use. Digestive diseases and sciences, 53(4), pp.954-963.
Kumar, A., Nawabpet, P. and Raju, G.S.K., (2022). Panchagavya as Soil Conditioner: Ancient Traditional Knowledge for Sustainable Agriculture. Journal of Experimental Agriculture International, 44(11), 181-186.


