1Department of Computer Science, Tarakeswar Degree College, Hooghly, West Bengal, India
2Department of physiology, Medical College and Hospital, Kolkata, West Bengal, India
3Department of Nutrition, AKPC Mahavidyalaya, Bengai, Hooghly, West Bengal, India
4Queen`s University, 99 University Ave, Kingston ON K7L 3N6
5Department of Computer Science, The University of Burdwan, West Bengal, India.
6Parasitology and Microbiology Research Laboratory, Department of Zoology, The University of Burdwan, West Bengal, India.
Corresponding author email: soumen.microbiology@gmail.com
Article Publishing History
Received: 09/04/2020
Accepted After Revision: 28/05/2020
Bacteria are important disease causing agents in human and subhuman vertebrates. This study demonstrated a unique method to identify the bacteria up to their species level on the basis of key biochemical properties. A soft-computing-based Intelligent Neural Network (INN) method has been proposed to classify the pathogenic bacteria such that Escherichia coli, Enterobacter aerogenes, Klebsiella pneumonia, Shigella dysenteriae, Salmonella typhimurium, Proteus vulgaris, Pseudomonas aeruginosa, Alcaligenes faecalis, Staphylococcus aureus, Lactococcus lactis, Micrococcus luteus, Corynebacterium xerosis, and Bacillus cereus with maximum accuracy. The methodology comprised two layered fully connected neurons to build the INN model and it is trained by using input vector having binary encoded values, from biochemical properties of taken bacterial set. Common biochemical properties of pathogenic bacteria have been taken for this study and a tabulation has been prepared by encoding the properties into zero(‘0’) for which species does not react positive and the properties encode into one(‘1’) where the species react positive. In this study the proposed INN method has been trained through forward and backward propagation by optimizing the Sum Squared Error of the network. To get the best outcome, the proposed method has been trained four times by varying epochs and learning rate, and the Sum Squared Error(SSE) has been reduced each time and finally it reached in ‘0.001165’ which is the best regarding classification of these bacterial set. This study would help laboratory users about the pathogenic bacteria to identify them in a fast and easy way.
Taxonomy, pathogenic, Soft-computing, SGD, INN, Sum Squared Error
Khamaru A, Saha I, Bandyopadhyay R, Chakraborty A. N, Karforma S, Chatterjee S. Taxonomy of Medically Important Bacterial Species Through Intelligent Neural Network: A Soft-Computing Based Approach. Biosc.Biotech.Res.Comm. 2020;13(2).
Khamaru A, Saha I, Bandyopadhyay R, Chakraborty A. N, Karforma S, Chatterjee S. Taxonomy of Medically Important Bacterial Species Through Intelligent Neural Network: A Soft-Computing Based Approach. Biosc.Biotech.Res.Comm. 2020;13(2). Available from: https://bit.ly/3aXcvVH
Copyright © Khamaru et al., This is an open access article distributed under the terms of the Creative Commons Attribution License (CC-BY) https://creativecommns.org/licenses/by/4.0/, which permits unrestricted use distribution and reproduction in any medium, provide the original author and source are credited.
INTRODUCTION
Nowadays it is challenging to determine the exact bacterial strain causing a specific, unidentified disease. However, the precise identification of bacteria is imperative in the treatment of resultant diseases. Bacteria are omnipresent, inhabiting water, soil, plants and also in other living organisms. The intestinal tracts of humans and animals are particularly rich, including pathogenic bacteria, such as those belonging to the Enterobacteriaceae family; most of the members of this family are gram-negative, non-spore forming facultatively anaerobes (Brisse et al., 2017; David and Paterson, 2012).
In this paper we have focused on disease causing bacterial species such as Escherichia coli, Enterobacter aerogenes, Klebsiella pneumonia, Shigella dysenteriae, Salmonella typhimurium, Proteus vulgaris, Pseudomonas aeruginosa, Alcaligenes faecalis, Staphylococcus aureus, Lactococcus lactis, Micrococcus luteus, Corynebacterium xerosis, and Bacillus cereus and their proper classification using the Intelligent Neural Network.
Escherichia coli is a free-living pathogen which is mainly found in human intestines. It is responsible for many infections, such as those of the urinary tract and colon as reported by (Meredith and Ulrich, 2013). Within the E. coli species, the strain E.0157 is particularly dangerous due to its production of the Shiga toxin. Within more vulnerable populations, such as children and the elderly, infection by this strain can be devastating, causing renal failure, hemolytic anemias, organ failure, and mental disequilibrium as reported by (Davis, 2018).
Less severe but still significantly, E.coli 0157 can cause chronic diarrhoea and dehydration. Chronic diarrhoea and dehydration from E. coli develop through the ingestion of contaminated food. The identification of the offending strain becomes possible when it is isolated from an infected patient’s stool or blood. Enterobacter aerogenes, also known as Aerobacter aerogenes, is a gram-negative, rod-shaped bacterium of the Enterobacteriaceae family. It is ubiquitous in the environment and generally present in fresh water, soil, vegetables and the excrement of human and animals.
It most commonly infects vulnerable populations such as young children, the elderly, and patients in the ICU. E. aerogenes is transmitted as nosocomial infections, as well as within the community. It can infect numerous organ systems such as the central nervous system, colon, urinary tract, skin, and blood. E. aerogenes can be the cause of meningitis if the bacteria can reach into the cerebrospinal fluid (Khan, 2004; McGrath ,2017).
Despite its low motility rate (10.2%), hospital environments can also induce its growth and transmission. However, E. aerogenes often acts in association with other pathogens. It is generally easily treatable with antimicrobials.
Klebsiella pneumonia is another common pathogen that warrants discussion. It is a gram-negative, non-motile, and rod-shaped bacterium belonging to the Enterobacteriaceae family (Amako et al., 1988). Often hospital-acquired, it is the cause of numerous infections from pneumonias to urinary tract and intra-abdominal infections (Amako et al.,1988; Ashurst and Dawson, 2019).
It is also a frequent respiratory pathogen which causes pneumonia by some symptoms like hemoptysis, and high fever. The prevalence of K. pneumoniae in community-acquired pneumonia has been reduced; however its prevalence in causing other pathology, such as liver abscess and meningitis, has been increased. K. pneumoniae is usually amenable to routine antimicrobial treatment. However, it is notable that antibiotic-resistant strains are emerging, which may present a challenge to treatment (Ashurst and Dawson 2019).
Shigella dysenteriae is one of the main species of Shigella. Being a member of the Enterobacteriaceae family, it is a small, gram-negative, non-motile, non-spore forming bacterium (Niyogi, 2005). It often resides in contaminated food and water, passing between hosts via either ingestion of the above or through fecal-oral contact. Houseflies have also been found to be a notable medium for infection by S. dysenteriae. It is responsible for bloody diarrhea, fever, abdominal pain, and stomach cramps in human (Keusch et al., 2011).
S. dysenteriae can be life-threatening, potentially causing Shigellosis, which is especially prevalent in developing countries. The Shiga toxin generated by this bacterium is responsible for Shigellosis. Beyond the acute danger of Shigellosis, less mild but chronic effects include malabsorption, growth retardation, and recurrent infections, which are especially prominent in already malnourished children (Niyogi, 2005). Salmonella typhimurium is gram-negative, rod shaped bacterium generally found in the human intestinal lumen (Keusch et al., 2011; Patel and McCormick, 2014).
Its medical significance lies in its ability to cause typhoid fever (Gart et al., 2016 Ashurst and Woodbury, 2019).
Proteus vulgaris is also gram-negative, rod shaped bacterium, which is widely spread in the environment and mainly found in the human gastrointestinal tract, water, soil, and excrement. As opportunistic pathogen, it is the 3rd most common cause of hospital-acquired infections (Bahashwan and Shafey, 2013). P.vulgaris is a motile, chemoheterotroph, and non-spore forming genera with various modes of transmission (Bahashwan and Shafey, 2013; Broeck and Herter, 1911). It has urease enzymes that allow it to produce ammonia in bulk, which is responsible for treating bladder and kidney stones. P. vulgaris is often amenable to treatment with antibiotics and bio-field treatment (Broeck and Herter, 1911; Braton et al., 2015).
Pseudomonas aeruginosa is a gram-negative, rod-shaped opportunistic pathogen and a member of the Enterobacteriaceae family. It is the cause of chronic infections in the urinary tract, skin (burn or surgical wounds), and respiratory tract of humans (Dzvova et al., 2016). It is ubiquitous in nature under distinct environmental conditions. P.aeruginosa is widespread in nature, soil, and water. This bacterium is well-grown in between 250C to 370C temperature (Bai F et al., 2015). P.aeruginosa is considered as hospital-acquired infections, particularly in immunocompromised patients. Antibiotics are required to treat this bacterial infection (Bai F et al., 2015; Cafasso, 2016).
Alcaligenes faecalis is a gram-negative, rod shaped motile, and aerobic pathogen (Mohammadizad and Salehizadeh, 2009), generally found in soil and water, as well as laboratory settings. A.faecalis is an opportunistic pathogen, which most often infects through liquid-based media such as nebulizers, respirators, and lavage of fluids (Mohammadizad and Salehizadeh , 2009). It can be found in the blood, urine, and feces. A more specific but notable manifestation in humans is the corneal ulcer. In addition to fluids, A. faecalis has been known to contaminate medical devices as well.
S.aureus is a gram-positive, facultatively anaerobic, naturally unencapsulated coccus. S.aureus is responsible for infections in newborn infants, breastfeeding women, and those with chronic disease such as vascular disease, lung disease, cancer and diabetes (Foster, 1996). This bacterium is a notable cause of sepsis through bacteremia. Infections due to S.aureus can be transmitted from one person to another always. Antibiotics are imperative in treating any infections caused by S.aureus.
Lactococcus lactis is a gram-positive, lactic acid-producing bacterium massively used in cheese, buttermilk, pickled vegetable productions and other types of fermented products. L.lactis is occasionally pathogenic, with few cases of disease reported. L.lactis is considered as pathogenic bacteria; some cases of diseases have been reported like infective endocarditis in adults and in children (Georgountzos et al. 2018).
It was first found in green plants and had the capability to grow on various sugars. L.lactis can boost the immune system to strive to recover allergies, hypertension, and it has more beneficial effects on the skin, and IBD (In et al., 2017).
Micrococcus luteus is a gram-positive coccus bacterium, about 0.5-3.5 micrometer in size, that can be found in water, soil, dust, and skin. M. luteus thrives in high oxygen environments, contributing to its prevalence in the respiratory tract and mucosal linings of the upper pharynx of humans (Rakhashiya et al., 2015). M.luteus can also grow in other environments, such as those with high salt concentrations, or little water at a 370C temperature (Krishnaveni and Umadevi, 2013).
Corynebacterium xerosis is a gram-positive, rod-shaped pathogen that can be found in skin and mucous membranes of humans. C.xerosis has been shown in normal skin flora, conjunctivas, and nasopharynx and most recently it has been isolated from vaginal swabs. Over the last two decades it has been recognized as a significant pathogen to humans, known to cause septicemia, endocarditis, pneumonia, osteomyelitis, septic arthritis, meningitis and ventriculitis especially in surgical patients or vulnerable populations like the immunocompromised or neonates (Cattani et al., 2000).
Bacillus cereus is a gram-positive, spore-forming, rod-shaped, motile pathogen. B.cereus is generally found in soil as well as a variety of foods. Its spore-forming characteristics help it to survive in severe environments where temperature is high or low. Its toxin production is the cause of gastrointestinal symptoms such as vomiting and diarrhea (Gherardi, 2016). In addition to its role in GI pathology, B.cereus can infect some surgical equipments and cardiac equipments in situ such as pacemakers, prosthetic mitral valves (Gherardi, 2016).
These thirteen bacteria display a broad diversity in significance within human pathology, highlighting the value of precise identification, often through phenotypic and biochemical characterization. Currently, bacterial taxonomy is based on the polyphasic approach comprising phenotypic, biochemical, serotypic and molecular methods. Soft computation is a new dimension in the field of bacterial taxonomy; here neural networking has been used as a soft computing-based classifier for the taxonomy of medically important pathogenic bacteria. An Intelligent Neural Network (INN) is a mathematical model for information processing, which is inspired by the neural system of the human brain (Hassankashi, 2019).
Now, INNs have become part of a technological revolution which have been focused on concrete decision making such as face recognition, gene identification and so on. We are now in the digital universe where many complex operations are executed through programmable intelligent electronic devices. Neural network technology is extensively used in both practical fields such as voice recognition and finger print recognition and academic fields such as pattern matching, data analytics, and programmable pathological equipment development and so on. In this study, INNs have been applied as a soft computing-based classifier in the taxonomy of pathogenic bacterial species.
We reviewed some significant literature regarding the classification of bacteria and their species. Huang et al. in their study applied, supervised and unsupervised machine learning techniques have been applied in eighteen classes of bacteria. Most of the bacterial species are known as disease-causing agents of humans. Three different algorithms were used in to classify species based on colony morphology. The supervised classification method comprising traditional and special Convolution Neural Network(CNN) has been applied. On the other side, Auto encoder has been included in the unsupervised method to classify bacteria. The deep neural network architecture of seven layers has been used and accuracy up to 90% has been achieved for distinguishing each bacterial species.
Manzoor et al.(2014) have introduced a new method for classification of bacterial strains of the same species. In this study, they applied the method on 40 bacterial strains, which are generally acquired in hospital. The proposed method based on a neural networks algorithm was applied to detect mutations and genetic variations in the bacterial strains. Principal Component Analysis (PCA), Discriminant Function Analysis (DFA) and the Probabilistic Neural Networks (PNN) algorithms were explored in a sensory perception system for recognition of a pattern in bacterial species from contaminated water (Carrillo and Durán, 2019).
Most early studies explored the role of NN in the analysis of colony morphology to detect mutations and genetic variation in bacteria. In the above studies all the authors have done their works through pattern recognition techniques, which focused on extracting the features applying on image processing from bacterial colony images. However, there is scanty literature available for the more advanced use of NN using biochemical properties to characterize and identify bacterial species. During the present study, the Neural Network based Intelligent Neural Network method has been explored in the taxonomy of medically important bacterial species through their fundamental biochemical properties.
MATERIAL AND METHODS
In this section, it has been discussed how the Intelligent Neural Network (INN) has been designed to classify different bacterial species belonging to their respective common pathogenic group. The cultural and biochemical characteristics of unknown organisms are separated and these characteristics have been encoded into a binary value. These values are fed into a trained INN to precisely identify and classify organisms.
In this method, two-layered intelligent neural networks have been used and the training process has been done through both forward and backward propagation. In the forward pass, the following function has been used where the synaptic weights of neurons would be unchanged throughout the network, and the signal would be computed for each neuron (Haykin, 1999; Padhy and Simon, 2015). The function generated output for neuron j as
![]()
Where, n was the number of iterations and ∂j(n) was the calculated output at the local field of neuron j, defined as
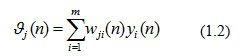
In the above equation, m implied a total number of input signals from neuron i to neuron j with wji (n) connecting synaptic weights and was the output of neuron i to the input in neuron j. Many hidden layers were used to design the proposed network model where the first hidden neuron in the network indicated by the value 1 in m in equation (1.2) and referred as yj(n)= yj(n)was output of input node xi(n) where i was indicated the first input terminal of the INN. The activation function in equation (1.1) was a Logistic function defined as:
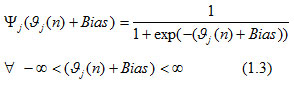
This was a sigmoidal nonlinearity function that generated an output on the induced local field of neuron j and Bias. The output amplitude of neuron j lied inside the range for a nonlinearity activation function (Haykin, 1999). This function was used at each level in all neurons with the same Bias to compute the output. The backward pass was the next phase of this method, where the output from the last neuron j was compared with the actual desired output and measured the amount of error signal generated by neuron j at iteration n, defined as
![]()
where dj(n) was the desired output and yj(n) was the actual output from neuron j at iteration n.
Cost function: In general, cost function depends on error energy generated by the network model at each level of outcome. The error energy was calculated for neuron j is
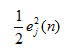
ξ(n) and the total error energy comes from all neurons in the output layer of the network as (Haykin, 1999; Padhy and Simon, 2015) defined by:.
 (2.2)
(2.2)
In this equation, C refers to the set of all neurons in the last layer of the network. In this study, 13 patterns have been used to train this network model, so that the average squared error energy will be calculated by: 1) adding all the error energy obtained in the equation (2.2) and then 2) dividing that sum by the number of patterns used in the network (Haykin, 1999), which is defined as:
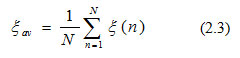
where N implies the number of patterns, which is 13 in this equation and ξav is the cost function used to measure the learning performance. Next, the error obtained from each neuron’s outcome was reduced, which was possible by updating weight vectors through the backward pass in each iteration. Weight vectors in this INN model were updated by the stochastic gradient descent (SGD) technique (Haykin, 1999). The small correction in synaptic weight vector wji(n)
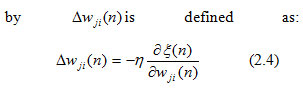
where n is the learning-rate assigned in this network as 0.001 and is unchanged throughout the training process. The back-propagation algorithm uses partial derivatives to correct weight vectors at all layers in the network, which happens through the chain rule of calculus, shown as:
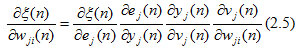
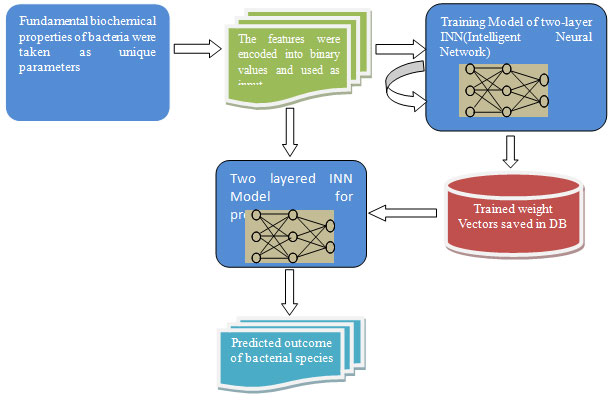
2.2 Flowchart of the Proposed Method
after encoding the features into 0 and 1 as in the table-3.
Step 2: Random values (-1 to +1) were initialized in two weight vectors: W1 and W2 respectively, and learning rate was initialized in ɳ with number of epochs ρ. The desired output was initialized in Y,
Step3: Two-layered INN model was trained as follows
While i in 1 to Number of Epoch ρ:
OH:=sigmoid(Dot product of (W1,X)+1)
OY:=sigmoid(Dot product of (W2,OH)+1)
Proposed Algorithm
Step 1: Bacterial features were extracted from Table-2 and used as the input vector X
OE:=OY – Y
Dlo3:=(2-OY)*OY*OE
Tx3:= Transpose of (OH)
Dld3:=Dot product of (Dlo3,Tx3)
W2:=W2 – ɳ*Dld3
Twx2:=Transpose of (W2)
Dld2:= Dot product of (Twx1,Dlo3)
Dlo1:=(2 – OH)*OH*Dld2
Tx:=Transpose of(X)
Dld1:= Dot product of (Dlo1,Tx)
W1:=W1 – ɳ*Dld1
i:=i+1
Step4: At the end of the training process, the weight vectors W1 and W2 were saved in two different files DB1 and DB2 respectively.
Step5: To predict bacteria in the Table-2, a small set of features was applied from input vector X into trained INN model-
Lx:=Sub set of (X)
TW1:=Load data from (DB1)
TW2:=Load data from (DB2)
XO:= sigmoid(Dot product of (TW1,Lx)+1)
PO:= sigmoid(Dot product of (TW2,XO)+1)
PO, the predicted output, that showed individual probability as a result of all labeled bacteria.
Biochemical characteristics of organisms taken into present consideration: trains of Escherichia coli were shown as positive or negative for curd, gas, and reduction in litmus milk reaction and only positive for methyl red and negative for H2S production, urease activity and citrate reaction. Strains of Enterobacter aerogenes were positive for acid reaction and citrate utilization and negative for H2S production, methyl red reaction and urease activity.
Strains of Klebsiella pneumonia were positive for acid and gas production in litmus milk reaction, methyl red test, urease activity and citrate utilization and negative for H2S production. Strains of Shigella dysenteriae were negative for urease activity, citrate utilization and H2S production and positive for methyl red test and alkaline reaction. Strains of Salmonella typhimurium are positive for alkaline reaction, methyl red, H2S production and citrate utilization and negative for urease activity. Strains of Proteus vulgaris are positive for all features taken into consideration in this study such as litmus milk reaction, methyl red, H2S production, and urease activity but are positive or negative for citrate utilization. Strains of Pseudomonas aeruginosa are positive for rapid peptonization reaction and citrate utilization and negative for methyl red, H2S production, and urease activity. Strains of Alcaligenes faecalis are positive for alkaline reaction and negative for methyl red, H2S production, and urease activity but are positive or negative for citrate utilization. Strains of Staphylococcus aureus are negative for citrate test, H2S production, and urease activity and positive for acid reaction and methyl red. Strains of Lactococcus lactis are positive for litmus milk reaction and methyl red and negative for citrate test, H2S production, and urease activity. Strains of Micrococcus luteus are positive for litmus milk reaction and urease activity and negative for all remaining features such as H2S production, methyl red, and citrate test. Strains of Corynebacterium xerosis are positive for litmus milk reaction and negative for H2S production, methyl red, urease activity and citrate test. Strains of Bacillus cereus are negative for H2S production, methyl red test, urease activity and citrate utilization and positive for peptonization reaction (Cappuccino and Sherman, 2009).
Data Representation: In this study, all biochemical characteristics of thirteen bacterial species have been encoded by binary value 0 and 1. Each biochemical characteristic of a particular species shows the reaction on use of some specified reagents (Cappuccino and Sherman 2014). The biochemical reaction has been shown in the form of symbols -, +, and ± and also some meaningful words which are used to indicate the proper reactions. These symbolic representations of reactions were encoded in 0 and 1 where, 1 indicate positive and 0 indicate negative effect of biochemical activity.
Table 1: An overview of thirteen disease causing organism and their cultural characteristics [Source: Cappuccino and Sherman, 2014].
| Bacterial Species | Shape/Gram Stain( + / – ) | Agar Slant Cultural features | Responsible for |
| E.coli | Rod /- | White, moist, glistening growth | Infection in the urinary tract, traveler’s diarrhea, renal failure, anemia, dehydration; organ failure, and mental disequilibrium |
| E. aerogenes | Rod /- | Abundant, thick, white, glistening growth | Nosocomial infections and meningitis |
| K.pneumoniae | Rod/ – | Slimy, white, somewhat translucent, raised growth | Pneumonia, liver abscess, and meningitis |
| S.dysenteriae | Rod/ – | Thin, even, grayish growth | Shigellosis, diarrhea, fever, abdominal pain, and stomach cramps |
| S.typhimurium | Rod /- | Thin, even, grayish growth | Fever, acute intestinal inflammation, and diarrhea |
| P.vulgaris | Rod /- | Thin, blue-gray, spreading growth | Hospital-acquired infections |
| P.aeruginosa | Rod /- | Abundant, thin, white growth, with medium turning green | Hospital acquired severe infections |
| A. faecalis | Rod*/ – | Thin, white, spreading, viscous growth | Corneal ulcer in human eye |
| S.aureus | Cocci/ + | Abundant, opaque, golden growth | Vascular disease, cancer, lung disease, and diabetes |
| L.lactis | Cocci/+ | Thin, even growth | Infective endocarditis in adults and in children |
| M.luteus | Cocci /+ | Soft, smooth, yellow growth | Impaired resistance in patients and colonizing the surface of heart valves |
| C.xerosis | Rod/ + | Grayish, granular, limited growth | Septicemia, peritonitis, endocarditis, pleura pneumonia, osteomyelitis, septic arthritis, meningitis |
| B.cereus | Rod /+ | Abundant, opaque, white waxy growth | Diarrhea, nausea, and vomiting |
Table 2: Disease causing organisms and their biochemical reactions [Source: Cappuccino and Sherman, 2014.]
| Bacterial Species | Litmus Milk Reaction | H2S Production | MR reaction | Urease activity | Citrate |
| E.coli | Acid, curd±, reduction± | – | + | – | – |
| E. aerogenes | Acid | – | – | – | + |
| K.pneumoniae | Acid, gas, curd± | – | ± | + | + |
| S.dysenteriae | Alkaline | – | + | – | – |
| S.typhimurium | Alkaline | + | + | – | + |
| P.vulgaris | Alkaline | + | + | + | ± |
| P.aeruginosa | Rapid peptonization | – | – | – | + |
| A. faecalis | Alkaline | – | – | – | ± |
| S.aureus | Acid, reduction ± | – | + | – | – |
| L.lactis | Acid, rapid reduction with curd | – | + | – | – |
| M.luteus | Alkaline | – | – | + | – |
| C.xerosis | Alkaline | – | – | – | – |
| B.cereus | Peptonization | – | – | – | – |
Table 3: Input vector of binary value [1= positive, 0=negative] for training to the proposed INN model of thirteen pathogenic bacteria.
| Bacterial Species | Litmus Milk Reaction | H2S Production | MR reaction | Urease activity | Citrate | ||||||||||||||
| Acid | curd± | reduction± | gas | gas± | Alkaline | Rapid peptonization | Rapid reduction with curd | Peptonization | – | + | – | + | ± | – | + | – | + | ± | |
| E.coli | 1 | 1 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 1 | 0 | 1 | 0 | 1 | 0 | 0 |
| E. aerogenes | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 1 | 0 | 0 | 1 | 0 | 0 | 1 | 0 |
| K.pneumoniae | 1 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | 1 | 0 | 1 | 0 |
| S.dysenteriae | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 1 | 0 | 1 | 0 | 1 | 0 | 0 |
| S.typhimurium | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 1 | 0 | 1 | 0 | 0 | 1 | 0 |
| P.vulgaris | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 1 | 0 | 0 | 1 | 0 | 0 | 1 |
| P.aeruginosa | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 1 | 0 | 1 | 0 | 0 | 1 | 0 | 0 | 1 | 0 |
| A. faecalis | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 1 |
| S.aureus | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 1 | 0 | 1 | 0 | 1 | 0 | 0 |
| L.lactis | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 1 | 0 | 0 | 1 | 0 | 1 | 0 | 1 | 0 | 0 |
| M.luteus | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | 1 | 0 | 0 | 0 | 1 | 1 | 0 | 0 |
| C.xerosis | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | 1 | 0 | 0 | 1 | 0 | 1 | 0 | 0 |
| B.cereus | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 1 | 0 | 1 | 0 | 0 | 1 | 0 | 1 | 0 | 0 |
RESULTS AND DISCUSSION
The INN algorithm has been proposed for the purpose of training and predicting the established disease-causing bacterial species separately in the classification model. The model is feasible on the Table-1 of thirteen bacterial species which are responsible for causing human disease. This classification model has been used to distinguish all
these bacterial species efficiently. A database of thirteen bacterial species has been taken from Bergey’s manual of Systematic Bacteriology where the biochemical characteristics like Litmus Milk Reaction, H2S production, MR reaction, Urease activity and Citrate utilization are taken as input for the INN. Biochemical properties are decoded into the vector. The vector is then translated into binary values and applied to the INN for training.
Table 4: Comparison of four training processes through INN model.
| Training No. | Model | Learning rate | Epochs | SSE |
| 1 | INN | 0.01 | 10000 | 0.180615 |
| 2 | INN | 0.01 | 100000 | 0.011125 |
| 3 | INN | 0.015 | 10000 | 0.010414 |
| 4 | INN | 0.015 | 100000 | 0.001165 |
The proposed model has been trained in four different ways by varying epochs and learning rate.
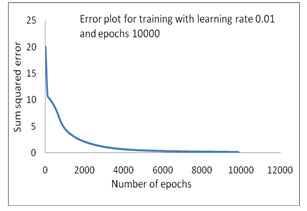
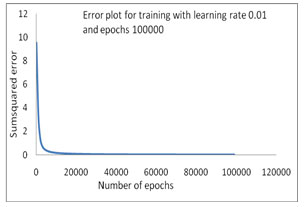
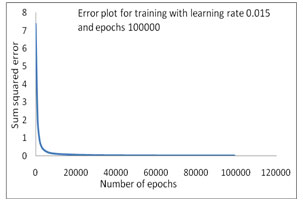
The error convergence plot of bacterial taxonomy has been accomplished by using INN model (Fig 1-4). First training has been plotted and shown in Figure 1, where as the Sum Squared Error (SSE) is 0.180615 on applied learning rate 0.01 and 10000 epochs but in comparison with same learning rate and the epochs has been increased up to 100000 to train this model in 2nd time where SSE is 0.011125 a better training, that as shown in figure 2. In the 3rd training process which is mentioned in the Table-4 where learning rate 0.015 has been initiated with 100000 epochs to train the INN model and the sum squared error generated is 0.010414 which is shown in Figure 3.
| Training No. | Model | Learning rate | Epochs | SSE |
| 1 | INN | 0.01 | 10000 | 0.180615 |
| 2 | INN | 0.01 | 100000 | 0.011125 |
| 3 | INN | 0.015 | 10000 | 0.010414 |
| 4 | INN | 0.015 | 100000 | 0.001165 |
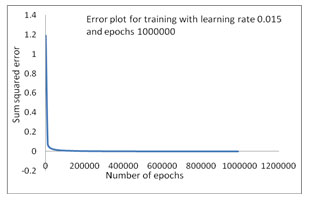
It has been indicated that the increase of epochs can reduce the SSE, so that the network has been trained 4th time which as in the Table-4 by initiating 1000000 epochs with learning rate 0.015 and has generated the SSE 0.001165 which is depicted in Figure 4. Moreover, the lower error-trained (SSE 0.001165) INN model has performed best regarding the recognition of bacterial species with an accuracy of 99.95%. We used an Intel core i5 processor with 2.6GHz frequency to train our proposed model. In every training period, we have noticed the time consumption that has increased due to the increment of epochs. 10000 epochs, with a learning rate of 0.0 1 took 2 seconds; 100000 epochs with a learning rate of 0.015 took 9 seconds and thus the others combinations of epochs and learning rate has been taken less time. The training period comprising 1000000 epochs with a learning rate of 0.015 took 105 seconds. The difference between seconds and minutes in the practical setting is an acceptable sacrifice if it will significantly improve accuracy. Our focus was first and foremost on the outcome of accuracy, as contrast to the time taken to reach it. The prediction accuracy was calculated by the relation of confusion matrix as
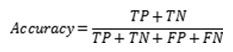
Where TN and TP refer to true negative and true positive respectively and their sum was 12.9928 for applying learning rate 0.015 with 1000000 epochs; on the other side FP and FN refer to false positive and false negative respectively and their sum was 0.2606 for same learning rate and epochs. The overall prediction accuracy was 98.03%. It has been calculated on the confusion matrix and it is considered as best predictions result. The error convergence plots of four training processes has been figured out from 1- 4 respectively, according to applied learning rate and epochs into the INN-
Figure 1: Error Convergence plot with SSE 0.180615
Figure 2: Error Convergence plot with SSE 0.011125
Figure 3: Error Convergence plot with SSE .010414
Figure 4: Error Convergence plot with SSE0.001165
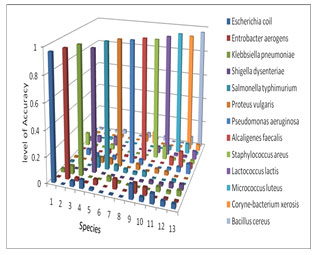
To compare the accuracy of the actual outcome with the predicted outcome, two plots have been depicted. A 3-D plot of Figure 5 shows that the accuracy in overall predictions is 97.05% when the model is trained with 10000 epochs and 0.01 learning rate. Raising the number of epochs and using optimum learning rate marginally increased accuracy each training period.
Figure 5: Accuracy plot of predicted bacterial species, where 10000 epochs with 0.01 learning rate have been used to train the INN.
Eventually, the best result was achieved by using 1000000 epochs with 0.015 learning rate, where as accuracy was 98.03%, as shown in figure 6. In the peruse of two figures 5 and 6 respectively, a clear perception is achieved that the plane of Figure 5 exhibits some small bars respect to the tall bars when 1st train cycle is used for separating of thirteen bacterial species. The average prediction rate was 97.05% for all thirteen bacterial species where SSE was 0.180615. In that case, the small altitude bars have been obtained which has indicated that no bacterial species will be predicted properly but in case of Figure 6 the average prediction rate was 98.03% for all bacterial species where SSE was 0.001165 and there was no small altitude bars in the plane which indicated that all the bacterial species have been predicted properly with high accuracy.
Figure 6: Accuracy plot of predicted bacterial species, where 1000000 epochs with 0.015 learning rate have been used to train the INN.
CONCLUSION
A two-layered INN model has been proposed for distinguishing disease-causing bacteria species based on biochemical properties such as Litmus Milk Reaction, H2S production, MR Reaction, Urease Activity, and Citrate utilization test. This model would help the researchers to identify the bacterial pathogens having fundamental biochemical properties. The proposed soft computing-based INN model is best suited for the classification of these medically important bacterial species.
Conflicts of Interests: The authors have no conflict of interest to declare.
ACKNOWLEDGEMENT
The authors wish to acknowledge the Zoology and Computer Science Department of The University of for the research assistance provided through the Support for laboratory equipments.
REFERENCES
Amako K, Meno Y, Takade A (1988). Fine Structures of the Capsules of Klebsiella pneumoniae and Escherichia coli K1. American Society for Microbiology Journals (Journal of Bacteriology).170( 10): 4960–4962.
Ashurst A and Dawson A (2019). Klebsiella pneumonia. Stat Pearls, https://www.ncbi.nlm.nih.gov/books/ NBK519004/ , Online.
Ashurst A, Truong J, Woodbury B. (2019). Salmonella typhi. StatPearls , https://www.ncbi.nlm.nih.gov/books/ NBK519002/, online.
Bahashwan SA, Shafey HM (2013). Antimicrobial resistance patterns of Proteus isolates from clinical specimens, September Edition, European Scientific Journal: 9(27).
Bai F, Jin Y, Jin S and Wu W (2015). Pseudomonas aeruginosa, Molecular Medical Microbiology. Second Edition. Science Direct. 2:753-767.
Branton A, Nayak G, Trivedi D, Trivedi M K (2015). Phenotyping and genotyping characterization of Proteus vulgaris after biofield treatment. Science Publishing Group.3(6):66-73.
Cohen J, Powderly W, Opal S (2010). Infectious diseases. 3rd edn. Elsevier, New York. P-2070.
Broeck TC and Herter CA (1911). A biochemical study of Proteus vulgaris. Journal of Biological Chemistry. 9: 491.
Cafasso J (2016). Pseudomonas Infections.www.healthline.com/health /pseudomonas-infections. online.
Cappuccino J and Sherman N (2009). Microbiology A Laboratory Manual. 7th Edition.Pearson, India.1-528. Cappuccino J and Sherman N. (2014).
Microbiology A Laboratory Manual. 10th Edition, Pearson, India: 216-217.
Carrillo J and Durán C (2019). Fast identification of Bacteria for Quality Control of Drinking Water through A Static Headspace Sampler Coupled to a Sensory Perception System. Biosensors. 9(1):23.
Cattani S, Berardi A, Venturelli C. (2000). Sepsis caused by Corynebacterium xerosis in neonatology: report of a clinic case. Acta Biomed Ateneo Parmense. 71( 1) :777-80.
David L and Paterson (2012). Infections due to other members of the Enterobacteriaceae, including management of multidrug-resistant strains. 24th Edition, Elsevier. 2:1874-1877.
Davis PC (2018). E.coli 0157: H7 infection early symptoms, treatment, and prevention. https://www.medicinenet.com/ e_coli__0157h7/article.htm, Online.
Dzvova N, Hamood AN, Hamood JA, Kruczek C (2016). Chapter six-In vitro analysis of Pseudomonas aeruginosa virulence using conditions that mimic the environment at specific infection sites. Progress in Molecular Biology and Translational Science. 142: 151-191.
Foster T (1996). Medical Microbiology, 4th Edition, The University of Texas Medical Branch, Galveston (TX)
Gart E V, Suchodolski J S, Welsh T H Jr. (2016). Salmonella typhimurium and multidirectional communication in the gut. Front Microbiol. 7: 1827.
Georgountzos G, Grivokostopoulos C, Michopoulos C (2018). Infective Endocarditis in a young adult due to Lactococcus lactis: A case report and review of the literature. Case Reports in Medicine. Article ID 5091456: 4.
Gherardi G (2016). The Diverse Faces of Bacillus cereus. Ist edn. Elsevier. 93-106.
Hassankashi M (2019). Neural Network. www.codeproject.com; Articles: 28, online.
Haykin S (1999). Neural Networks A Comprehensive Foundation. Second Edition, Pearson, India: 178-266.
Huang L and Wu T (2018). Novel neural network application for bacterial colony classification. Springer: 22.
Song AA., In, L.L.A., Lim, S.H.E. et al. Erratum to: A review on Lactococcus lactis: from food to factory. Microb Cell Fact 16, 139 (2017). https://doi.org/10.1186/s12934-017-0754-1
Joo H S, Hirai M, Shoda M (2007). Improvement in ammonium removal efficiency in wastewater treatment by mixed culture of Alcaligenes faecalis no. 4 and L1, J Biosci Bioeng. 103(1):66-73.
Keusch GT, Kopecko DJ, Mohammed A S (2011). Shigellosis, Third Edition. Elsevier. Chapter-18. 137-144.
Khan AF(2004). Meningitis due to Enterobacter aerogenes subsequent to resection of an acoustic neuroma and abdominal fat graft to the mastoid. Brazilian Society of Infectious Diseases. 8(5).
Krishnaveni M and Umadevi K (2013). Antibacterial activity of pigment produced from Micrococcus luteus KF532949. Elsevier. 4(3): 149-152.
Licitra G (2013). Etymologia: Staphylococcus, Centers for Disease Control and Prevention. 19(9):1553.
Manzoor S, Moncayo S, Navarro-Villoslada F (2014). Rapid identification and discrimination of bacterial strains by laser induced breakdown spectroscopy and neural networks. Elsevier. 121: 65-70.
McCormick BA and Patel S (2014). Mucosal Inflammatory Response to Salmonella typhimurium Infection. Front Immunol. 5:311.
McGrath S (2017). Enterobacter aerogenes & Disease. https://healthyliving.azcentral.com/enterobacter- aerogenes-disease-12320900.html. Online.
Meredith TA and Ulrich JN (2013). Retina. Fifth Edition.Science Direct:2019-2039.
Mohammadizad S and Salehizadeh H. (2009). Microbial enhanced oil recovery using biosurfactant produced by Alcaligenes faecalis. Irian Journal for Biotechhnology. 3: 7(4):216-223.
Niyogi SK (2005). Shigellosis. The Journal of Microbiology. 43:133-143.
Padhy NP and Simon SP (2015). Soft computing with MATLAB Programming. First Edition. Oxford University Press. India: 118-152.
Rakhashiya PM, Patel PP, Thaker VS. (2015). Whole genome sequences and annotation of Micrococcus luteus SUBG006, a novel phytopathogen of mango. Elsevier: 6:10-11.