1Ministry of Health and Medical Education, NIMAD, Tehran, Iran.
2Department of Parasitology and Mycology, Cellular and Molecular Research Center, Qazvin University of Medical Sciences, Qazvin, Iran.
3Department of Medical Entomology & Vector Control, School of Public Health, Tehran University of Medical Sciences, Tehran, Iran.
4Department of Parasitology and Mycology, Tehran University of Medical Sciences, Tehran, Iran.
Article Publishing History
Received: 24/08/2016
Accepted After Revision: 24/09/2016
Cutaneous leishmaniasis (CL) is a worldwide public health and a social problem in many developing countries. The main objective of this study was to investigate the relationship between interleukin 4 (IL-4) gene promoter polymorphisms and leishmanial infected people in endemic focus of zoonotic cutaneous leishmaniasis (ZCL), in north east of Iran. In order to determine of polymorphism of interleukin 4 (IL-4) among patients with different clinical symptoms, fifty human blood samples were prepared. All cases were classified into five groups including: cases with sever and ≤ 2 lesions, cases with ≥ 2 lesions, cases with treatment duration of less than two months, cases with treatment duration of more than two months and healthy samples with no signs. All 50 human samples were tested by PCR and followed using restriction enzyme of Eco47I. Our study revealed that Eco47I RE could cut the PCR product to an 18 bp and 177 bp if the SNP was C. Among the 50 PCR products we found 4 samples (8%) with CT allele and also 1 sample(2%) with TT allele. The rest of samples (90%) were CC. In conclusion, there was a significant difference on frequencies of three alleles (CC, CT, TT) in studied group (÷2=56.4; df=2 ; p=0.000).
Snp, Interleukin 4, Polymorphisms, Cutaneous Leishmaniasis, Iran
Rafizadeh S, Saraei M, Abai MR, Mohebali M, Bakhshi H, Rassi Y. Relationship Between Interleukin 4 Gene Promoter Polymorphisms and Cutaneous Leishmaniasis Cases in North Eastern Iran. Biosc.Biotech.Res.Comm. 2016;9(3).
Rafizadeh S, Saraei M, Abai MR, Mohebali M, Bakhshi H, Rassi Y. Relationship Between Interleukin 4 Gene Promoter Polymorphisms and Cutaneous Leishmaniasis Cases in North Eastern Iran. Biosc.Biotech.Res.Comm. 2016;9(3). Available from: https://bit.ly/2BrCGEw
Introduction
Leishmaniasis is a parasitic disease with a wide spectrum of clinical manifestations ranging from a self-healing skin lesion to lethal form of visceral disease. According to WHO estimate the prevalence of leishmaniasis is 12 million with 0.9–1.6 million new cases each year. Leishmaniasis occurs in 98 countries worldwide, and 350 million people are at risk of contracting the disease. Zoonotic Cutaneous Leishmaniasis (ZCL) is the most common form of leishmaniasis and about one-third of cases are reported from the Americas, the Mediterranean basin, and Western Asia from the Middle East to Central Asia. Countries such as Afghanistan, Algeria, Colombia, Brazil, Iran, Syria, Ethiopia, North Sudan, Costa Rica and Peru, which together account for 70 to 75% of global estimated ZCL cases. It is shown that the distribution of ZCL in central and south Asian countries such as Kazakhstan, Kyrgyzstan, Turkmenistan, Uzbekistan, Tajikistan, Iran, Pakistan, Afghanistan, southern Mongolia, and north-western China overlaps with the presence of great gerbils, the main reservoir of ZCL in Iran (Sosnina, 1979; Mallon, 1985; Yaghoobi-Ershadi and Javadian, 1996, Strelkova et al., 2001; Molur et al., 2005; Abai et al., 2010; Smith et al., 2010; Tashbaev and Mustafaev, 2010; Oshaghi et al., 2011; Alvar et al., 2012; WHO 2012 and Bakhshi et al., 2014).
Leishmaniasis clinical manifestation depends upon Leishmania species and host genetic background which governs generation of type of immune response (Mohamed et al., 2003; Salhi and Rodrigues, 2008; ; Sakthianandeswaren et al., 2009 and 2010; Castellucci et al., 2012; Moravej et al., 2012, Bakhshi et al., 2013 and 2014).There is correlation between the type of immune response induced and cytokines production and outcome of the diseases. It is well known that in murine model of L. major infection dichotomy of Th1/Th2 immune response determines the outcome of the disease (Reiner and Locksley, 1995). In leishmaniasis usually cytokines such as IFN-©, IL-4, IL-5, IL-10, TNF-〈, TNF-®, etc in the level of protein and genes are titrated to assess the severity and outcome of CL due to L. major (Habibi et al., 2001; Mahmoodi et al., 2003; Kamali-Sarvestani et al., 2006; Sakthianandeswaren et al., 2009; Salhi and Rodrigues, 2008).
IL-4 plays an important role in various biological activities including immune response development and its polymorphisms is reported from different populations (Kamali-Sarvestani et al., 2006). Correlations between IL-4 polymorphisms and different disorders such as visceral leishmaniasis (VL), CL, and leprosy are reported (Mohamed et al., 2003; Kamali-Sarvestani et al., 2006; Yang et al., 2011).There is a report that in human IFN-c + 874 A>T polymorphisms influences the progression of the disease towards chronic CL while IL-4 -590 C>T polymorphism increases the risk of developing CL (Kamali-Sarvestani et al., 2006).
In 2014, a PCR-based assay was developed to amplify IL-4 promoter gene to possibly define IL-4 promoter gene polymorphism in R. opimus populations with a range of Leishmania infection and symptoms collected from different foci of the central, north and northeast regions of Iran. The results showed that the designed primers amplify 689 bp of the promoter gene and Sequence analysis of the promoter gene revealed five polymorphic sites assembly six haplotypes among the gerbil populations (Bakhshi et al., 2014).
This is important that the five polymorphisms cause different outcome phenotypes following infection with L. major in R. opimus specimens. In the current study it was proposed to investigate the relationship between interleukin 4 (IL-4) gene promoter polymorphisms and leishmanial infected people in endemic focus of cutaneous leishmaniasis in north east of Iran.
Material And Methods
This study were carried out in Esfarayen County during 2014-2015. This county is located in North Khorasan Province, north-east of Iran (Fig.1). At the 2006 census its population was 51,321, in 13,376 families. Esfarayen is one of the focal points for residence of Aryan tribes after entering into Iran. Basis on combination of clinical and parasitological criteria, fifty cases were selected from three villages of Charborj(CH), Kalateh Reza(KR) and Hossein Abad (HA). All samples were classified into five groups (each group=10 cases) including: cases with sever and ≤ 2 lesions, cases with ≥ 2 lesions, cases with treatment duration of less than two months , cases with treatment duration of more than two months and healthy samples with no lesions or scars. Blood samples were prepared from all selected groups.
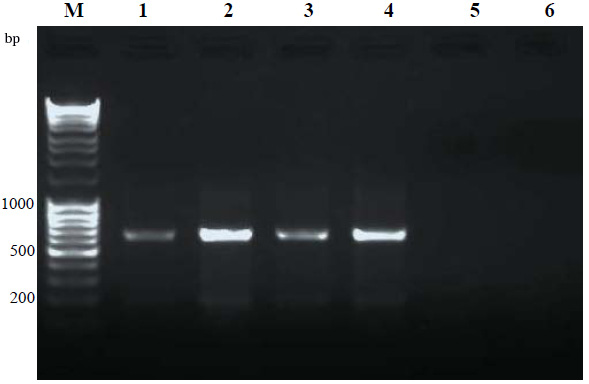 |
Figure 1: The map of North Khorasan Province, North-East of Iran, Esfarayen district has been located in south of the province |
Two hundred μL of the frozen blood samples were thawed and treated with proteinase K, and then the genomic DNA was extracted using the G-spin Tissue Spin Kit (G-spin, South Korea) according to the manufacturer’s instructions. The sequences of the designed primers were as follow: F-: 5’-TGG GGA AAG ATA GAG TAA TA-3’ and R-: 5´- TAA ACT TGG GAG AAC ATG GT-3’ (Kamali-Sarvestani et al., 2006).
The primers amplify a fragment with size of 195 bp. Amplification was performed on an Eppendorf thermal cycler. A touch-down PCR thermal program was carried out with the following profile: 94, 53 and 72 degrees centigrade for 50 seconds respectively. PCR products were then used for RFLP by Eco47I Restriction Enzyme and then the amplified products were monitored by electrophoresis in 2.5% agarose gel and ethidium bromide staining.
The expected band was purified from the gel by gel extraction kit (Bioneer, South Korea). Sequencing was performed using an ABI 3730 sequencer machine and Seqlab (Göttingen, Germany). The ambiguous sequences were corrected using Chromas program and the consensus sequences obtained using DNASTAR Lasergene (SEQMAN and EDITSEQ). The sequences were aligned using ClustalW (Chenna et al., 2003) to explore possible polymorphisms.
Results And Discussion
The aim of this investigation was to determine of promoter region of IL-4 gene and its correlation with people infected to Leishmania major. All samples were amplified and finally the PCR products were used to evaluation by the use of Eco47I restriction enzyme. The results revealed the presence of 3 genotypes including CC, CT and TT. The most prevalent genotype belonged to CC type (90%). Eight percent of samples (4 cases) had an genotype of CT and finally just one of the samples had a genotype of TT (2%). In the group of people with no lesions or scars we only observed CC genotype. Also we observed a TT genotype in the group of people which had been treated less than two months. The other samples out of this group had CC genotype. Also we observed 1/10 CT and 9/10 CC genotypes in the group which had been treated more than two months. We found 1/9 CT genotype and 9/10 CC genotypes in the group with severe lesions. Finally we found 1/10 CT genotype and 9/10 CC genotypes in the group belonged to samples wih small lesions, (Table 1).
| Table 1: The observed genotype and allele frequencies for interleukin gene promoter, among five selected group in Esfarayen county, North east of Iran, 2015 | |||||
| G1 N (%) | G2 N (%) | G3 N (%) | G4 N (%) | G5 N (%) | |
| IL-4 genotype | |||||
| CC | 8 (80) | 9 (90) | 9 (90) | 9 (90) | 10 (100) |
| CT | 2 (20) | 1 (10) | 0 (0) | 1 (10) | 0 (0) |
| TT | 0 (0) | 0 (0) | 1 (10) | 0 (0) | 0 (0) |
| IL-4 allele | |||||
| C | 18 (90) | 19 (95) | 18 (90) | 19 (95) | 20 (5) |
| T | 2 (10) | 1 (5) | 2 (10) | 1 (5) | 0 (0) |
| G1(Acute lesions > 2), G2(Mild lesions ≤2 ), G3(Treatment time ≤ 2 month)
G4 (2 month < Treatment time ≤ 6month) and G5 (Healthy cases without lesion or scar) |
|||||
Our study showed that Eco47I RE could cut the PCR product to an 18 bp and 177 bp if the SNP was C (Fig 2.). A single nucleotide polymorphisms (SNPs) in the IL-4 promoter gene of the infected people were identified which might be related to the pathogenesis and clinical outcome of leishmaniasis. Of course the impact of this SNP on the outcome of the disease should be tested independently, because all of the mutations in cytokine genes may not have similar influence on the clinical outcomes of leishmaniasis. In human, it is reported that TNF-á -308 G–>A and TNF-â +252 G–>A gene polymorphisms showed no effect on the disease but IFN- ã+874 A–>T polymorphism influences the progression of the disease towards chronic CL while IL-4 -590 C–>T polymorphism might increase the risk of developing CL (Kamali-Sarvestani et al., 2006).
The results of this study were analyzed by SPSS-Version 18 and X2 (Non parametric) soft wares. Due to this analyze, the prevalence of CC, CT and TT alleles among 5 mentioned groups revealed that these 3 allele prevalence in the selected population has significant differences (÷2=56.4; df=2 ; p=0.000); but we could not find a significant difference in prevalence of CC, CT and TT alleles in three selected villages of this investigation ÷2=5.6; df=2 ; p=0.062). Also we could not observed any significant difference in CC and CT alleles among the 5 groups (÷2=1.3; df=1 ; p=0.248); but the prevalence of CC, CT and TT alleles in 4 groups (cases with less than two lesions, Treatment period < 2 months, Treatment period > 2 months, Healthy without lesions or scars) had a significant difference (÷2=4.5; df=1 ; p=0.035).
In a study performed in human visceral leishmaniasis, it was shown that IL-4 polymorphism but not IL-9 influences VL incidence and clinical phenotypes due to L. donovani infection (Mohamed et al., 2003). In contrast, IFNGR1 polymorphism was linked and associated to post Kala-azar dermal leishmaniasis (PKDL) but not VL. The authors concluded that polymorphism in a type 2 cytokine gene influences underlying susceptibility to VL, whereas IFNGR1 is specifically related to susceptibility to PKDL. In a study performed in CL due to L. braziliensis, it was shown that a SNP in macrophage inhibitory factor (MIF -173C) favored CL infection and disease progression (De Jesus Fernandes Covas et al., 2013).
In another study it was shown that a SNP in IL-1â (-511 C/T) is possibly a key player determining the severity of the disease in diffuse (DCL) patients infected with L. mexicana (Fernández-Figueroa et al., 2012). Also it was shown that IL-6 -174 G/C promoter polymorphism influences susceptibility to mucosal but not localized CL (Castellucci et al., 2006).
In another investigation conducted on R. opimus specimens in Iran, six IL-4 gene promoter region haplotypes among sequenced specimens observed. Sequence analysis of these samples revealed five single nucleotide polymorphisms (SNPs) assembly six haplotypes among the gerbil populations .Four out of five SNPs (80%) were of transition type (A-G or T-C). The inter population genetic variations ranges from zero to 1% .There was no significant correlation between the haplotypes and the geographical origins or subspecies of the great gerbils but some specific geographical haplotype were seen (Bakhshi et al., 2014).
Conclusion
In this study, presence of three different genotypes of CC, TT, CT among leishmaniasis cases was confirmed the polymorphism in the promoter of IL-4. Also, the high frequency of CC genotype (90%) was observed compared with the other two genotypes. More importantly, healthy individuals with no ulcers were observed with CC genotype, that inconsistent with the results of the other investigators in Iran. Therefore, it seems that the endemicity of the disease (Hypo, Meso and Hyper endemic) has a significant impact on results. Finally, further studies are needed to discover SNPs impressing the disease in the human populations and also rodents particularly in R. opimus, which is the main reservoir host of ZCL. These data may lead to develop novel control strategy against ZCL.
Acknowledgement
The authors gratefully thank Esfarayen Health Center, Northern Khorassan province, north east of Iran for its field assistance. This study was financially supported by the School of Public Health, Tehran University of Medical Sciences (Project No.24217) and Qazvin University of Medical Sciences.
References
Abai MR, Oshaghi MA, Tajedin L, Rassi Y, Akhavan AA (2012). Geographical distribution and ecological features of the great gerbil subspecies in the main zoonotic cutaneous leishmaniasis foci in Iran. Asian Pacific Journal of Tropical Medicine, 3: 800-803.
Alvar J, Vélez ID, Bern C, Herrero M, Desjeux P, Cano J, Jannin J, den Boer M (2012).Leishmaniasis worldwide and global estimates of its incidence. PLoS One , 7: e35671.
Bakhshi H, Borhani N, Mohebali M, Khamesipour A, Abai MR, Hajjaran H , Tajedin L , Rassi Y, Akhavan A A , Mohtarami F, Oshaghi M A (2014).Interleukin 4 (IL-4) gene promoter polymorphisms in Rhombomys opimus, the main reservoir of zoonotic cutaneous leishmaniasis. Cytokine , 65: 1-3
Castellucci L, Jamieson S E, Almeida L, Oliveira J, Guimarães L H , Lessa M , Lago E , de Jesus A R, Carvalho EM, Blackwell J M (2012).Wound healing genes and susceptibility to cutaneous leishmaniasis in Brazil. Infection, Genetics and Evolution, 12:1102-1110
Castellucci L , Menezes E, Oliveira J, Magalhães A, Guimarães LH, Lessa M, Ribeiro S , Reale J, Noronha E F, Wilson ME, Duggal P , Beaty T H , Jeronimo S, Jamieson S E , Bales A , Blackwell J M , de Jesus A R, Carvalho E M (2006). IL6− 174 G/C promoter polymorphism influences susceptibility to mucosal but not localized cutaneous leishmaniasis in Brazil. Journal of Infectious Diseases ,194:519-527.
Chenna R, Sugawara H, Koike T, Lopez R , Gibson TJ, Higgins DG,Thompson J D (2003). Multiple sequence alignment with the Clustal series of programs. Nucleic Acids Research 31:3497-3500.
De Jesus Fernandes Covas C, Cardoso C C, Gomes-Silva A, Oliveira J R S, Da-Cruz AM , Moraes M O (2012). Candidate gene case-control and functional study shows macrophage inhibitory factor (MIF) polymorphism is associated with cutaneous leishmaniasis. Cytokine , 61:168-172.
Fernández-Figueroa E A , Rangel-Escareño C, Espinosa-Mateos V, Carrillo-Sánchez K, Salaiza-Suazo N, Carrada-Figueroa G,March-Mifsut S, Becker I (2012). Disease Severity in Patients Infected with Leishmania mexicana Relates to IL-1â. PLoS Neglected Tropical Diseases ,6:e1533.
Habibi GR,Khamesipour A, McMaster W, Mahboudi F (2001). Cytokine Gene Expression in Healing and Non Healing Cases of Cutaneous Leishmaniasis in Response to In vitro Stimulation with Recombinant gp63 Using Semi‐Quantitative RT–PCR. Scandinavian Journal of Immunology,54: 414-420.
Kamali-Sarvestani E, Rasouli M , Mortazavi H, Gharesi-Fard, B. (2006). Cytokine gene polymorphisms and susceptibility to cutaneous leishmaniasis in Iranian patients. Cytokine, 35: 159-165.
Mallon D(1985).The mammals of the Mongolian People’s Republic. Mammal review , 15:71-102.
Mohamed H, Ibrahim M, Miller E, Peacock C, Khalil E, Cordell H J,Howson
J M, El Hassan A M, Bereir R E, Blackwell J M (2003).Genetic susceptibility to visceral leishmaniasis in The Sudan: linkage and association with IL4 and IFNGR1. Genes and Immunity, 4:351-355.
Mahmoodi M, Khamesipour A, Dowlati, Rafati S, Momeni A, Emamjomeh M, Hejazi H, Modabber F(2003).Immune response measured in human volunteers vaccinated with autoclaved Leishmania major vaccine mixed with low dose of BCG. Clinical & Experimental Immunology, 134: 303-308.
Molur S, Srinivasulu C, Srinivasulu B, Walker S, Nameer P, Ravikumar L ( 2005). Status of Non-Volant Small Mammals. Conservation Assessment and Management Plan (CAMP) Workshop Report, Coimbatore, India, 2005.
Moravej A , Rasouli M, Kalani M, Asaei S, Kiany S, Najafipour S , Koohpayeh A , Abdollahi A (2012) . IL-1â (− 511T/C) gene polymorphism not IL-1â (+ 3953T/C) and LT-á (+ 252A/G) gene variants confers susceptibility to visceral leishmaniasis. Molecular Biology Reports, 39: 6907-6914.
Oshaghi M, Rassi Y,Tajedin L, Abai M R, Akhavan A A, Enayati A, Mohtarami F (2011).Mitochondrial DNA diversity in the populations of great gerbils, Rhombomys opimus, the main reservoir of cutaneous leishmaniasis. Acta Tropica , 119: 165-171.
Reiner S L, Locksley R M (1995). The regulation of immunity to Leishmania major. Annual Review of Immunology,13:151-177.
Smith A T, Xie Y, Hoffmann R S, Lunde D, MacKinnon J, Wilson DE (2010). A guide to the mammals of China, Princeton University Press.,p.213–214.
Sakthianandeswaren A, Foote S J, Handman E (2009). The role of host genetics in leishmaniasis. Trends in Parasitology, 25: 383-391.
Sakthianandeswaren A , Curtis J M, Elso C, Kumar B, Baldwin T M, Lopaticki S, Kedzierski L, Smyth G K, Foote S J, Handman E (2010). Fine mapping of Leishmania major susceptibility locus lmr2 and evidence of a role for Fli1 in disease and wound healing. Infection and Immunity,78: 2734-2744.
Salhi A, Rodrigues Jr V, Santoro F, Dessein H, Romano A, Castellano L R, Sertorio M , Rafati S, Chevillard C, Prata A, Alcaïs A, Argiro L, Dessein A (2008). Immunological and genetic evidence for a crucial role of IL-10 in cutaneous lesions in humans infected with Leishmania braziliensis. J Immunol , 180: 6139-6148.
Sosnina E F ( 1979). The lice of the gerbils of Tajikistan. Vshi peschanok Tadzhikistana , 13: 29-35.
Strelkova M, Eliseev LN, Ponirovsky EN, Dergacheva TI, Annacharyeva DK, Erokhin PI, Evans DA (2001). Mixed leishmanial infections in Rhombomys opimus: A key to the persistence of Leishmania major from one transmission season to the next. Annals of Tropical Medicine and Parasitology , 95:811-819.
Tashbaev N and Mustafaev K (2010). Current epidemiology of zoonotic cutaneous leishmaniasis in the Republic of Uzbekistan. Meditsinskaia Parazitologiia i Parazitarnye Bolezni , 4: 34-36.
World Health Organization (2012).http://www.who.int/leishmaniasis/burden/en/> (accessed January 2012).
Yaghoobi E and M R Javadian (1996). Epidemiological study of reservoir hosts in an endemic area of zoonotic cutaneous leishmaniasis in Iran. Bulletin of the World Health Organization,74:587-590.
Yang D, Song H, Xu W, Long H, Shi C, Jing SongW, Pei B (2011) Interleukin 4-590T/C polymorphism and susceptibility to leprosy. Genetic Testing and Molecular Biomarkers , 15: 877-881.