Department of Post Graduate Studies and Research in Microbiology, Gulbarga
University, Kalaburagi, Karnataka, India.
Corresponding author email: drvandanarathods@gmail.com
Article Publishing History
Received: 15/08/2022
Accepted After Revision: 25/09/2022
Keratinases from Aneurinibacillus aneurinilyticus are capable of degrading keratinous proteins. Salt precipitation and diethylaminoethyly determined the purification and characterization of the enzyme. Ion-exchange and gel permeation chromatography, and SDS-PAGE. Physicochemical factors like pH, temperature, metal ions, enzyme inhibitors and substrate. To study Km and Vmax various concentrations of keratin were used for the activity of enzyme. Gel permeation chromatography with 20.84-fold purification. 203.87 U/mg specific activity showed 34KDa between 14 to 31KDa in SDS-PAGE. The number was stable at pH 7.0-9.0 400-500C, and optimum at pH 9.0 and 500C.
Further stimulated by Mg2+, Ca2+, K+, Fe2+, Zn2+, Mn2+, and Na2+ inhibited by Cu2+, Co2+ and Hg2+. Ethylene diamine tetra acetic acid with the highest stimulatory effect was inhibited by Di-isopropyl fluoro phosphatase and phenyl methyl sulfonyl fluoride. Enzyme was stable with Tween-60, TritonX-100 and TritonX-114 declined with ß-mercaptoethanol. It hydrolyzed several keratinous substrates as keratin and casein were 100 and 85.47% utilized with Km=3mM, Vmax =249µmol/ml/min. Xerophytic endophytes are treasure houses as they tolerate biotic and abiotic stress, are stable at high temperatures and pH are selected, such keratinases can be used in leather processing and detergent industries.
Aneurinibacillus Aneurinilyticus, Characterization, Keratinase, Keratin, Purification.
Sujata S, Vandana R, Dattu S, Ravi M, Krishna R, Narmada S, Roopa N, Shajji D. Purification and Characterization of Keratinase from Aneurinibacillus aneurinilyticus Isolated from a Xerophytic Plant Opuntia ficus-indica. Biosc.Biotech.Res.Comm. 2022;15(2).
Sujata S, Vandana R, Dattu S, Ravi M, Krishna R, Narmada S, Roopa N, Shajji D . Purification and Characterization of Keratinase from Aneurinibacillus aneurinilyticus Isolated from a Xerophytic Plant Opuntia ficus-indica. Biosc.Biotech.Res.Comm. 2022;15(2). Available from: <a href=”https://bit.ly/3eeLIvn“>https://bit.ly/3eeLIvn</a>
Copyright © This is an Open Access Article distributed under the Terms of the Creative Commons Attribution License (CC-BY). https://creativecommons.org/licenses/by/4.0/, which permits unrestricted use distribution and reproduction in any medium, provided the original author and sources are credited.
INTRODUCTION
The insoluble keratin which is highly stable is present in fur, feathers, beak, horns, nails, and hair of living animals (Onifade et al. 1998; Pandian et al. 2012). The length of Keratin fibre depends on their water-containing complex hydrates, which increases their length by 10-12% (Bhuyar et al. 2018). According to the sulfur content, hair, nail, hoof and also feathers are grouped as hard keratins where as callus and skin belong to soft keratins. Hard keratins from chicken feathers obtained from feather industries have high level of disulphide bonds, cross linked structures which are resistant and insoluble, having hydrophobic interactions, (Annapurna et al. 1996). Chicken feathers constitute 5-7% of keratin producing a large number of poultry wastes which decomposes slowly causing massive environmental concerns, (Ningthoujam et al. 2016; Almahasheer et al. 2022).
Feather keratin characteristically attributes to the presence of various amino acids (Kumar et al. 2011; Gupta and Singh 2014). Physical and chemical methods of chicken feather degradation consumes high energy, damaging the products (Zoccola et al. 2012; Lee et al. 2016; Holkar et al. 2016). Hence, an alternative method is suggested for converting keratinous wastes into economic, reusable, eco-friendly products by using such enzymes produced from vast microbial sources. Such enzymes are produced by various keratinolytic microorganisms. Keratinous substrates can be hydrolyzed by keratin degrading enzymes which have potential applications. Keratinases are produced by fungi, bacteria and actinomycetes (Onifade et al. 1998; Gupta and Ramnani 2006; Bhuyar et al. 2018; Almahasheer et al. 2022). Feather degradation by the enzyme keratinase produced by B. subtilis, B. cereus, and B. licheniformis has been reported (Thys et al. 2004; Almahasheer et al. 2022).
Keratinase are used in several biotechnological processes, such as removing hair and feather for the production of feather meal, clearing the obstructions in the sewage systems and dehairing process in the leather industry (Gupta et al. 2002). Other uses include production of biogas, biodegradable films, glues, prion hydrolysis, wastewater processing, recovery of silver from X-ray films, drug delivery in medicine cosmetics and additives (Patinvoh et al. 2016; Almahasheer et al. 2022).
This work highlights the characterization and purification of the enzyme keratinase from Aneurinibacillus aneurinilyticus isolated from an endophytic xerophyte (Opuntia–ficus indica). Several studies have been done in the past on keratinase production, purification and characterization by bacteria. However, this paper is one-of-its-kind that has isolated endophytic bacteria from xerophytes in drought regions. These enzymes from such bacteria can tolerate high temperatures and pH which can be used in various biotechnological industries. The keratin degrading protein i.e., keratinases has diversified application in green technology. Therefore, there is a search for an efficient bacterial strain by the researchers (Almahashree et al. 2022). The keratin from poultry waste is degraded by the purified enzyme keratinases from B. licheniformis dcs1, has the efficiency to enhance the nutritional quality of the waste keratin from poultry, (Liaqat et al 2022).
MATERIAL AND METHODS
Bacteria was grown in 1000 ml production media under submerged fermentation and fermenting broth at 10,000 rpm at 40C for 10 mins and it was centrifuged. Obtained supernatant was considered as a crude enzyme for purification purposes and at each step, enzyme activity was estimated. The crude enzyme from A. aneurinilyticus was partially purified further, ammonium sulphate precipitation was used at different saturation levels (10-80%) as per the standard chart with continuous stirring overnight under cold conditions to precipitate the desired protein.
Collected precipitate was centrifuged at 40C for 10 min., further protein pellet was dissolved in 20 mmol/L Tris-HCl buffer (pH8.0). Folin-Phenol reagent along with bovine serum albumin was used as standard for protein estimation. The protein sample was loaded into the activated dialysis membrane and sealed at both ends. The dialysis 150 membrane bag was suspended in 500mL (50 mM, pH 7.0) phosphate buffer was used. Experiment was carried out with four changes in the buffer (Kamble et al. 2020).
The dialysate obtained from the above step was concentrated using (DEAE-cellulose), chromatographic anion coloumn (1.5X30cm). Sodium chloride with different concentrations (pH-8.0) in 20mmol/L Tris HCl buffer was used to elute the sample at 6 ml/min flow rate for collecting the fractions (Cheng-Gang Cai et al. 2008). The concentrated enzyme was equilibrated with 100mM Tris-HCl buffer (pH8.0) and Sephadex G-100 (50cm X 1.7 Cm) to prepare for gel filtration. The eluted pure protein fractions were characterized at the rate of 1ml/min of the fraction collected using same buffer. 10% separating acryl amide gel and 5% stacking gel having 0.1% SDS were used to visualize protein bands through Coomassei Brilliant Blue R-250. The protein bands were then destained with acetic acid and methanol. Pure enzymes were used for characterization (Lammli 1970; Cheng-Gang Cai et al. 2008).
Bovine Serum Albumin 66KDa, Lysozyme 14.3kDa, Phosphorylase 97kDa, Ovalbumin 45kDa and Bovine Carbonic Anhydrase 31 kDa, were used as standard protein markers (Eun-Jan Jeong et al. 2010). Zymogram analysis was performed by using 2% keratin as substrate copolymerized with stacking gel of 4% and 10% resolving gel, the sample buffer and purified proteins together were loaded to polymerized gel. Tris-HCl buffer 0.01M having a pH of 9.0 Triton X-100 (v/v) 2.0% were subjected to electrophoresis, for about half an hour. To remove Triton X-100 the gel was washed with D. H2O and incubated at 370C for 30 min.
After incubating at pH-9.0 and 370C for 30mins in Tris-Cl, stained with Coomassie Brilliant Blue Dye R-250 for half an hour then de-stained, clear colourless zone was detected and considered as enzyme activity (Vermelho et al. 2009). For the characterization of purified keratinase, various physicochemical factors like temperature, pH, metal ions, enzyme inhibitors along with substrate were studied. Enzyme activity was noted using 0.1M acetate buffer (pH3-5), sodium phosphate buffer 0.1M (pH 6.0-8.0), glycine sodium hydroxide buffer (pH 9.0-12.0) with 0.1M Tris HCl buffer (pH 7.0-9.0) and temperature 10-800C were taken to study the effect of pH and temperature on enzyme activity. The residual activities were measured by studying the effect of temperature and pH on enzyme stability by incubating the enzyme solution at pH between 3.0-12.0 and temperature between 10-800C for 60 min (Murthy et al. 2019).
The effect of enzyme inhibitors such as Ethylene Diamine Tetra Acetic Acid (EDTA), Phenyl Methyl Sulfonyl Fluoride (PMSF), Dithiothreitol (DTT), Diisopropyl Fluoro Phosphate (DFP), Sodium Dodecyl Sulfate (SDS), β-mercaptoethanol, Triton X-114, Triton X-100, Tween-60, at 5mM (1%) along with Purified keratinase were pre-incubated in 100mM Glycine /NaOH buffer for 1h. The enzyme without inhibitor served as control and was taken as 100% activity (Ghasemi et al. 2012). Along with the effect of metal ions such as Cu2+, Co2+, Hg2+, Na2+, K+, Mg2+, Mn2+, Ca2+, Zn2+(1mM each) then incubating the purified fraction to determine the effect of metal ions.
Different substrates such as haemoglobin, bovine serum albumin, fibrin, gelatin, keratin, and casein (1% w/v) were used for determining the enzyme Substrate specificity. The enzyme was incubated for10 min in each of the substrates. The degree of substrate hydrolysis was analyzed by protease activity using standard assay conditions (Gupta and Singh 2014).
For measuring the enzyme activity various concentrations of keratin (1mM-10mM) dissolved in 0.05 M glycine NaOH buffer of pH 9.0 and incubation time of 30 min. under standard assay conditions were used for determining the Kinetic parameters Km, Vmax of purified keratinase by ploting Line weaver-Burk plot (1934).
RESULTS AND DISCUSSION
Enzyme mass production: Further the crude enzyme was used for purification steps.
Purification of crude enzyme: Salt precipitation, dialysis, Gel filtration, sephadex G-100, DEAE cellulose were used for keratinase Purification from A. aneurinilyticus. Over all it was observed that the specific activity was 203.87U/mg, yield of 25.43% with a fold increase of 20.84 (Table-1). Keratinases from P. vulgaris EMB-14 showed purification folds of 14.46 with specific activity of 74.74 U/mg and 8.27% yield as reported by (Dada et al. 2020). A 10-fold purification with 3.46% yield and specific activity of 297.5U/mg with recovery of 45% was and keratinase with 7.5-fold increases in a specific activity with 45% recovery was reported in the previous studies (Ire et al. 2017).
Keratinase with a 7.5-fold increase in a specific activity with 45% recovery (Adina et al. 2021). A similar result was observed with a single band at 26KDa both in zymogram and SDS PAGE with B. licheniformis H62 (Kazzaz et al. 2015). B. subtilis SCK6 showed 30.95KDa and B. amyloliquefaciens strain TCCC 11319 with 28KDa (Tian et al. 2019). Zhang et al. (2016) Han et al. (2012) also reported a nearer mol. wt. of 25KDa and 33KDa of keratinase from P. aeruginosa C11. The reports of various authors producing keratinase from Bacillus sps. correlate with our results (Tian et al. 2019; Adina et al. 2021).
Keratinase from A. aneurinilyticus and its molecular weight was determined by comparing the electrophoretic mobility of the marker proteins. Standard protein markers used were 97KDa (phosphorylase), 66KDa (BSA), 45KDa (Ovalbumin), and 31KDa (Bovine carbonic anhydrase) and 14.33KDa (Lysozyme). The mol. wt. of the purified enzyme was 34KDa when SDS PAGE was done (Fig.1a). Zymogram analysis showed a clear colourless zone against a dark blue background (Fig.1b). Similar results were observed with Kazzaz et al. (2015) revealing a single band at 26KDa both in zymogram and SDS PAGE with B. licheniformis H62.
B. subtilis SCK6 showed 30.95KDa and B. amyloliquefaciens strain TCCC 11319 with 28KDa (Tian et al. 2019, Zhang et al. (2016) and Han et al. (2012) also reported almost nearer mol. wt. of 25KDa and 33KDa of keratinase from P. aeruginosa C-11. Liaqat et al. (2022) also purified and characterized keratinase from B. licheniformis dcs1 from poultry waste processing and the keratinase which correlates our results, keratinase with mol. wt. of 34KDa.as reported by (Yong et al. 2020). Thus, showing similar reports of keratinase production with almost similar mol.Wt. from Bacillus sps (Yong et al. 2020; Liaqat et al. 2022).
Table 1. Steps of keratinase Purification from A. aneurinilyticus VRCS-4
| Steps of Purification | Activity (U/ml) | Protein (mg) | Specific activity (U/mg) | Fold of Purification | (%) Yield |
| Crude culture fluid | 38478 | 3931 | 9.78 | 1.0 | 100 |
| (NH4)2SO4
Precipitation (70%) |
27863 | 2453 | 11.35 | 1.16 | 74.41 |
| DEAE cellulose | 11602 | 143 | 81.13 | 8.29 | 30.15 |
| CM Sephadex G-100 Gel filtration | 9786 | 48 | 203.87 | 20.84 | 25.43 |
Figure.1 .a: SDS PAGE of keratinase (M: Marker protein), (1: Keratinase).
Figure.1 .b: Keratinase Zymogram

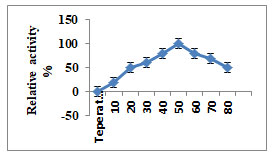
Characterization of Purified Enzyme: Maximum activity of purified keratinase enzyme was at 8.0 to10.0 pH, while moderate activity was between 5.0-11.0. Relative keratinase activity which was maximum (100%) was at pH 9.0 (Fig.2). To check the enzyme stability from A. aneurinilyticus it was pre incubated between the temperatures of 20-800C. It was observed that the enzyme was stable at 500C. Further, increase in the temperature to 800C, the relative activity was reduced to 40% (Fig.3).by increasing the temperature to 800C, revealing that the purified keratinase showed the optimum activity at pH9.0and temperature 500C (Kumar et al. 2021).
Similar results of optimum pH and temperature with B. megaterium were reported by Saibabu et al. (2012). Using the organism Chrysosporium indicum as reported by Kumar et al. (2021) it also showed maximum keratinase activity at temperature 500C and pH-10. Yong et al. (2020) reported optimal temperature, pH 550C and 10.0 by B. subtilis S1-4. When these results of keratinase production were compared with all the researchers it was observed that the pH ranged between 8.0 -9.0 and the temperature between 50-600C (Kumar et al. 2021).
Figure 2: Influence of pH on keratinase activity
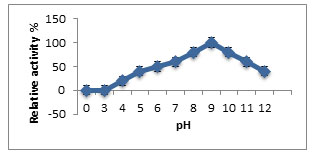
Figure 3: Influence of temperature on keratinase activity
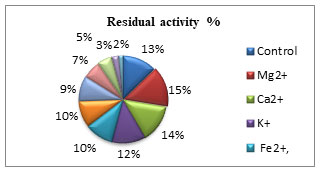
Keratinase activity on metal ions is presented in (Fig. 4). The metal ions, studied were Mg2+, Ca2+, K+, Fe2+, Zn2+, Mn2+, Na2+, Cu2+, Hg2+ and Co2+ at 1mM concentration. The percent residual activity was maximum with Mg2+, Ca2+, K+, Fe2+, Zn2+and Mn2+ while Cu2+, Co2+, and Hg2+ showed inhibitory action against keratinase from A. aneurinilyticus. Keratinase activity was inhibited by Hg2+, which signifies that Hg2+, might reduce the enzyme activity by binding to the SH-group present at the active site or Hg2+ may bind to the carboxylic group and also tryptophan residue to decrease the enzyme activity (Saibu et al. 2013; Ul–Haq 2020). While in case with A. aneurinilyticus some cations stimulated the keratinase activity. Saibabu et al. (2013) reported that the enzyme inhibition with Hg2+ is not due to the thiol group but is due to the tryptophan residue interaction or may be the carboxyl group present in the amino acid of that particular enzyme. The stimulatory effect of Fe2+, Ca2+and Mn2+ at 1mM increases the keratinase activity by acting as co-factors (Ul–Haq 2020).
Suntornsuk et al. (2005) reported that Ca2+ a divalent cat ions enhanced the enzyme activity of B. licheniformis FK-14, indicating the enzyme to be typical for serine protease. To maintain the enzyme structural confirmation or to stabilize the substrate binding and to form enzyme complex probably these metal ions may be acting as ion bridge. Keratinase from A. aneurinilyticus is highly activated by Mg2+, Ca2+ and Mn2+ proving that these metal ions confer for protection of enzyme against denaturation by heat performing a vital function in the maintenance of its active conformation (Kumar and Takagi 1999). Dada (2020) expressed that keratinase largely from gram-positive bacteria are mostly serine proteases as they possess two Ca2 + active sites thereby enhancing the enzymatic activity. The role of Ca2 + associated with the stability of the activated forms of the keratinases agrees with our results (Dada 2020; Ul–Haq 2020).
Figure 4: Effect of metal ions on keratinase activity
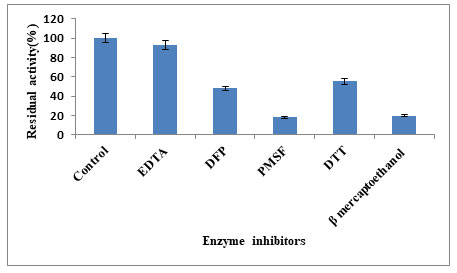
Keratinase was stimulated by EDTA, DTT, DFP while the most potent inhibitor was PMSF (Fig.5). When the surfactants results were compared the keratinase activity was inhibited (19.92%) to the maximum with β-mercaptoethanol and stimulated by tween-60 (84.11%), tritonX-100 (82.01%), tritonX-114 (58.65%) and SDS (32.46%) (Fig.6). Inhibition of the enzyme activity in presence of PMSF indicates the keratin belongs to serine group. This inhibition is due to covalent binding of PMSF to the residual serine and in activating it, there by blocking the sulphonate releasing hydrolytic sides for keratinolytic attack (Suntornsuk et al. 2005). The active site of protease was blocked by PMSF by sulfonating the essential serine residue, there by resulting in complete inhibition of protease activity (Jaouadi et al. 2013). However,reducing agents able to break the disulphide bonds in the substrate keratin releasing different hydrolytic sites for the keratinolytic attack (Dada 2020; Ul–Haq 2020).
Moreover 70% of reduction in enzyme activity was observed with β-mercaptoethanol. Xian et al. (2016) also presented 70% of enzyme inhibition with β-mercaptoethanol, while it was 55% with DTT. More than 50% of residual activity was with other nonionic surfactants. We evaluated the impact of inhibitors and surfactants on keratinase activity by A. aneurunilyticus. Enzyme activity was inhibited by PMSF, β-mercaptoethanol and stimulated by EDTA, while amongst the surfactant used is again stimulated by Tween-60 and anionic surfactant such as SDS inhibited them. Our results correlate with Bose et al. (2014) where PMSF completely inhibited the enzyme activity while nonionic and anionic were marginal stable. Stability of the keratinase enzyme from B. subtilis k-5 with SDS and Tween-80 while activity was inhibited by β-mercaptoethanol was reported by Singh et al. (2014). Rai and Mukharjee (2009) stated that the stable alkaline protease in presence of surfactants is highly desirable for industrial application (Ul–Haq 2020).
Figure 5: Effect of enzyme inhibitors on keratinase activity
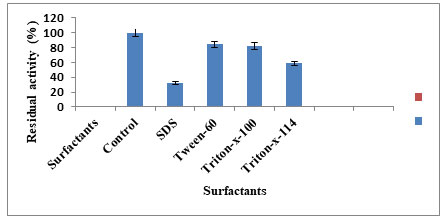
Figure 6: Effect of surfactant on enzyme activity
Keratinases from A. aneurunilyticus showed broad substrate specificity since it could hydrolyze keratin substrate and casein (85.47%), BSA (69.04%), gelatin (59.72%), haemoglobin (48.21%) and the least were fibrin (36.43%) (Table-2).
Moridshashi et al. (2020) reported maximum relative activity with feather keratin then was casein, and the least hydrolysed were keratin azure, gelatine and BSA. Prakash et al. (2010) reported high activity towards casein, followed by keratin, using B. halodurans PPKS-2. Gang et al. (2008) also revealed maximum keratinase activity by using casein, BSA, Feather meal and feather keratin with the organism B. subtilis KD-N2. When the relative activity of different substrates was compared it was observed that the substrates which were having more disulphide bonds could be easily hydrolysed than the substrates having less disulphide bonds, suggesting the solubility depends on the high percentage of disulphide bonds in the substrates (Moridshashi et al. 2020).
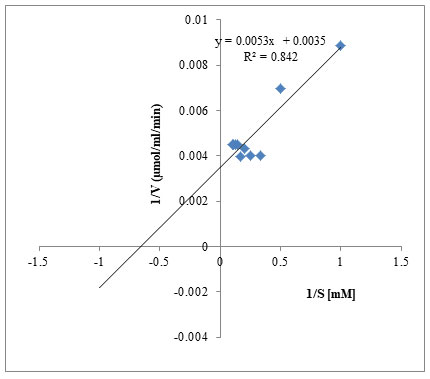
Different concentrations of the substrate keratin (1mM to10mM) were used to determine the Km and Vmax of purified keratinase. Km and Vmax values were determined by LB plot and were found to be 3mM-Km and 249µmol/mL/min Vmax respectively (Fig.7). Lower values of Km suggest high affinity towards the substrates indicating that the enzyme-substrate complex is tightly held before the substrate is converted to the product thus indicating that keratinase enzyme from A. aneurinilyticus has great affinity towards its substrate keratin. 1/S on the X-axis, 1/V on the Y-axis, a double reciprocal plot gave a straight line suggesting that our enzyme obeys the Michaelis Menton equation. Km being independent of enzyme concentration shows the characteristics of enzyme under defined temperature and pH condition (Moridshashi et al. 2020).
Singh (2014) and Srinivasan (2008) also reported a low Km value of 0.01µg/ml/min and Vmax of 1176mg/ml with B. subtillis K-1. Dada (2020) also reported Km and Vmax kinetic constants of 25.60mM and 74.46U/ml respectively, with B. licheniformis K-51. Hydrolysis efficiency of 7mg/ml for Km and 384.6U/mg of Vmax was observed when casein was the substrate, while it was 7.2mg/ml and 103mg/ml respectively when the substrate was keratin azure with Laeeyella sacchari (Dada et al. 2020). Moridshashi et al. (2020) also reported a low Km value of 8.74mg/ml and Vmax of 59.04U/ml/min with feather meal substrate using B. zhangzhouensis. The above-said report correlates with our data (Dada et al. 2020).
Table 2. Specificity of keratinase in the presence of different substrate
| Substrates | Enzyme activity (U/ml) | Relative activity (%) |
| Bovine serum albumin | 252.5 | 69.04 |
| Casein | 312.0 | 85.47 |
| Gelatin | 218.0 | 59.72 |
| Fibrin | 136.0 | 36.43 |
| Keratin | 365.0 | 100.00 |
| Haemoglobin | 176.0 | 48.21 |
Figure 7: Line Weaver-Burk plot (1/[S] V/S 1/ [V]) Km and Vmax
CONCLUSION
The findings of the present study showed that proteases of microbial origin are interesting, compared to plants or animal sources, as these enzymes from microbial origin possess all the features desired for biotechnological applications. Consequently, keratin can be transformed by keratinolytic microorganisms such as A. aneuinilyticus with low molecular weight of (34KDa). Generally, the molecular weight of bacterial keratinases varied between species and combination of various peptidases is required for keratin degradation. Thus, keratinase with low mol. wt. can be used in various biotechnological processes. Our studies of keratinase production from xerophytic endophytes will unravel the complex mechanism of keratinolysis and stability of the enzymes at high pH and temperatures which can be employed in various industries.
Ethics approval and consent to participate: Not applicable.
Funding: No funding.
Conflict of interests: Authors declare no conflict of interests to disclose.
ACKNOWLEDGEMENTS
The work was carried out in the Department of Microbiology, Gulbarga University, Kalaburgi, Karnataka, India, which is greatly acknowledged
REFERENCES
Adina DA, Ezeamagu CO, Akindele ST et al. (2021). Purification and Properties of Wickerhanomyces anomalus Keratinase and Its Prospective Application in Poultry Feed Industries. Fountain Journal of Natural and Applied Sciences. 10(1): 1-12.
Almahasheer AA, Mahmoud A, and El-Komy H, (2022). Novel Feather Degrading Keratinases from Bacillus cereus Group: Biochemical, Genetic and Bioinformatics Analysis, Microorganisms, Vol.10(1), PMC8781890, doi:10.3390/10010093.
Annapurna RA, Chandrababu NK, Samivelu N et al. (1996). Eco-friendly enzymatic dehairing using extracellular protease from Bacillus species isolate. J. Am. Leather Chem. Assoc., 91:115-119.
Balaji S, Kumar M, Karthikeyan R, et al. (2008). Purification and Characterization of an extracellular keratinase from a hornmeal-degrading Bacillus subtilis MTCC (9102). World J MicrobiolBiotechnol 24(1):2741-2745.
Bhuyar P, Zagade S, Revankar R et al. (2018). Isolation Characterization and Partial Purification of keratinase from keratinolytic Bacteria.Sch j Appl sci Res, Vol.1:16.
Bose A, Pathan S, Pathak K et al. (2014). Keratinolytic Protease Production by Bacillus amyloliquefaciens 6B Using Feather Meal as Substrate and Application of Feather Hydrolysate as Organic Nitrogen Input for Agricultural Soil Waste Biomass Valor, 5:595-605, DOI 10.1007/s12649-1013-9272-5.
Cheng SW, Hu HM, and Shen SW (1995). Production and Characterization of Keratinase of a Feather-degrading Bacillus licheniformis PWD-1. Biosci. Biotech. Biochem. 59(12):2239-2243.
Cheng-gang CAI CG, Chen JS, QI JJ, et al. (2008). Purification and Characterization of keratinase from a new Bacillus subtilis strain. Journal of Zhejiang University Science B 9(9):713-720.
Dada M and Wakil S (2020). Production, Purification and Characterization of Keratinase from Bacillus species Isolated from Poultry feather waste, Scientific Research Journal (SCIRJ), Vol.VII, Issue IV, 83. DOI: 10.31364/SCIRJ/v8.i4.2020.P0420767.
Ghasemi Y, Shahbazi M, Rasoul-Amini S, et al. (2012). Identification and Characterization of feather-degrading bacteria from keratin – rich wastes, Ann Microbiol, 62:737-744, DOI 10.1007/s 13213-011-0313-7.
Guleria S, Walia A, Chauhan A, et al. (2018). Purification and Characterization of detergdent stable alkaline protease from Bacillus amyloliquefaciens SP1 isolated from apple rhizosphere Journal of Basic Microbiology, Environment Health Techniques, DOI 10.1002/jobm.201500341.
Gupta R and Ramnani P (2006). Microbial keratinases and their prospective applications: an overview. Appl Microb Biotechnology 70:21-33. DOI : 10.1007/s00253-005-0239-8
Gupta R, Beg QK and Lorenz P (2002). Bacterial alkaline proteases: Molecular approaches and Industrial Applications. Appl Microbial Biotechnology. 59:15-32 DOI 10.1007/s00253-002-0975-y.
Gupta S and Singh R (2014). Hydrolyzing proficiency of keratinases in feather degradation. Indian J Microbial 54(4):466- 470.
Hamiche S, Mechri S, Khelouia L, et al. (2019). Purification and biochemical characterization of two keratinases from Bacillus amyloliquefaciens S13 isolated from marine brown alga Zonariatourne fortii with potential keratin- biodegradation and hide-unhairing activities. International Journal of Biological Macromolecules Vol.122 pages 758-769 https://doi.org/10.1016/j.ijbiomac.2018.10.174.
Han M, Luo W, Gu Q et al. (2012). Isolation and characterization of a keratinolytic protease from a feather-degrading bacterium Pseudomonas aeruginosa C11, African Journal of Microbiology Research Vol. 6(9), pp.2211-2221, DOI:10.5897/AJMR 11.921 ISSN 1996-0808.
Holkar C, Ananda J, Bhavasar JP et al. (2016). Acoustic Cavitation Assisted Alkaline Hydrolysis of wool Based Keratins to produce Organic Amendment Fertilizers. ACS Sustain Chem Eng. 4:2789-96. DOI:10.1021/acssuschemeng.6b00298.
Ire SF and Onyenama AC (2017). Purification and Some Properties of Keratinase from Bacillus licheniformis strain NBRC 14206. Journal of Applied Life Sciences International. 11(3):1-9, Article no. JALSI.33165.
Jaouadi NZ, Rekik H, Badis A et al. (2013). Biochemical and Molecular Characterization of a serine Keratinase from Brevibacillus brevis US575 with promising keratin- Biodegradation and hide- dehairing activities. PLOS ONE Vol. 8(10)e76722. https://doi.org/10.1371/journal.pone.0076722
Jeong E, Rhee M, Kim G et al. (2010). Purification and Characterization of a Keratinase from a Feather –degrading Bacterium, Bacillus sp. SH-517.J.Korean soc.Appl.Biol.Chem.53(1), 43-49 .
Kamble S, Jadhav A and Sonawane K (2020). An efficient de-hairing by Keratinase from Streptomyces badies var. Shashi, Eco. Env. and Cons. 26(1): 2020; pp. (193-198), ISSN 0971-765X.
Kazzaz AE, Feizi ZH, Guvenmez HK et al. (2015). Keratinolytic Protease Production and Characterization from Bacillus sps. Isolated from poultry wastes. International Journal of Applied Biology and Pharmaceutical Technology. Vol.6 (4).
Kumar CG and Takagi H (1999). Microbial alkaline protease: from bio industrial view point. Biotechnol.Adv.17:287-291.
Kumar EV, Srijana M, Chaitanya K et al. (2011). Biodegradation of poultry feathers by novel bacterial isolate Bacillus altitudinis GVC11.Indian J Biotechnology 10:502-507.
Kumar J and Mahal S (2021). Isolation of Chrysosporium indicum from poultry soil for keratinase enzyme, its purification and partial characterization. Journal of Applied and natural sciences 13(2):744-751, https://doi.org/10.31018/jans.v13i2.2609.
Laemmi UK (1970). Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227(5259):680-685.
Lee YS, Phang LY, Ahmad SA et al. (2016). Microwave – alkali treatment of chicken feathers for protein hydrolysate production. Waste Biomass Valorization.7:1-11.
Liaqat I, Ali S, Butt A, et al. (2022). Purification and Characterization of keratinase from Bacillus licheniformis dcs1for poultry Waste Processing. Journal of Oleo Science, 71(5) 693-700, DOI:10.5650/21426.
Lineweaver H and Burk D (1934). The Determination of enzyme constants. Journal of the American Chemical Society 56(3):658-666.
Moridshahi R, Bahreini M, Sharifmoghaddam M et al. (2020). Biochemical characterization of an alkaline surfactant – stable keratinase from a new keratinase producer, Bacillus zhangzhouensis, Extremophiles 24:63-704 https://doi.org/10.1007/s00792-01187-9.
Murthy VNY, Murulidhara NV and Mariswamy M (2019). Production and Purification of Keratinase Enzyme from Serratia sp. Isolated from Poultry Wastes, Journal of Applied Sciences, 19 (8):789-796, DOI:10.3923/jas.2019.789.796.
Ningthoujam DS, Devi LJ, Devi PJ et al. (2016). Optimization of Keratinase Production by Amycolatopsis sp. Strain MBRL 40 from a Limestone Habitat. J. Bioprocess Biotech. Vol. 6(5). 1000282. https://doi.org/10.4172/2155-9821.1000282.
Onifade AA, Al-Sane NA, Al-Musallam AA et al. (1998). A review: Potentials for biotechnological applications of keratin – degrading microorganisms and their enzymes for nutritional improvement of feathers and other keratins as livestock feed resources. Bioresource Technology, 66:1-11.
Pandian S, Sundaram J and Panchatcharam P (2012). Isolation, identification and characterization of feather degrading bacteria. European Journal of Experimental Biology, 2(1):274-282.
Patinvoh RJ, Lagerstedt EF, Lundin M et al. (2016). Biological Pretreatment of Chicken Feather and Biogas Production from Total Broth. Appl Biochem Biotechnol.180:1401-1414 DOI 10.1007/s12010-016-2175-8.
Paul T, Das A, Mandal A, et al. (2014). Biochemical and Structural Characterization of a Detergent Stable Alkaline Serine Keratinase from Paenibacillus woosongensis TKB2: A Potential Additive for Laundry Detergent. Waste Biomass Valor: 5:563-574 DOI :10.1007/s12649-013-9265-4.
Prakash P, Senigala K, Jayalakshmi et al. (2010). Purification and characterization of extreme alkaline, thermostable keratinase, and keratin disulfide reductase produced by Bacillus halodurans PPKS-2. Appl MicrobiolBiotechnol 87:625-633 DOI 10.1007/s00253-010-2499-1.
Rai S and Mukherjee AK (2009). Statistical optimization of production, purification and industrial application of a laundry detergent and organic solvent stable subtillisin like serine protease (Alzwiprase) from Bacillus subtillis DM-04. BiochemEng J 48:172-180.
Saibabu V, Niyonzima FN and Sunil SM (2013). Isolation, Partial purification and Characterization of Keratinase from Bacillus megaterium. International Research Journal of Biological Sciences Vol. 2(2),13-20.
Singh S, Gupta P, Sharma V et al. (2014). Multifarious potential applications of keratinase of Bacillus subtilis K-5. Biocatalysis and Biotransformation.32: 333–342, DOI: 10.3109/10242422.2014.978306.
Suntornsuk W, Tongjun J, Onnim P et al. (2005). Purification and characterization of keratinase from a thermo tolerant feather-degrading bacterium. World Journal of Microbiology and Biotechnology 21:1111-1117, DOI: 10.1007/s11274-005-0078-x.
Thys RC, Lucas FS, Riffel A et al. (2004). Characterization of a protease of a feather degrading Microbacterium species. Lett Appl Microbiol 39(2):181-186. DOI:10.1111/j.1472-765X.2004.01558.x.
Tian J, Xu Z, Long X et al. (2019). High – expression keratinase by Bacillus subtilis SCK6 for enzymatic dehairing of goat skins. International Journal of Biological Macromolecules 135 DOI: 10.1016/jbiomac.2019.05.131.
UlHaq I, and Akram F (2020). Keratinolytic enzyme –mediated biodegradation of recalcitrant poultry feathers waste by newly isolated Bacillus sp. NKSP-7 under submerged fermentation. Folia Microbiologican 65:823–834. https://doi.org/10.1007/s12223-020-00793-6.
Vermelho AB, Mazotto AM and Nogueira ACMD (2009). Identification of a Candida parapsilosis strain Producing Extracellular Serine Peptidase with Keratinolytic Activity. Mycopathologia. 169:57–65 DOI 10.1007/s11046-009-9231-7.
Yong B, Fei X, Shao H et al. (2020). Recombinant expression and biochemical characterization of a novel keratinase BsKER71 from feather degrading bacterium Bacillus subtilis S1-4. AMB Expr 10:9 https://doi.org/10.1186/s13568-019-0939-6.
Zhang RX, Gong JS, Zhan DD et al. (2016). A metallo-keratinase from a newly isolated Acinetobacter sp. R-1 with low collagenase activity and its biotechnological application potential in leather industry. Bioprocess BiosystEng 39:193–204 DOI 10.1007/s00449-015-1503-7.
Zoccola M, Aluigi A, Patrucco A et al. (2012). Microwave-assisted chemical-free hydrolysis of wool keratin. Text Res J. 82:2006–18 https://doi.org/10.1177/0040517512452948 .


