1Department of Clinical Embryology and Preimplantation Genetics, Chennai
Fertility Center and Research Institute, Chennai, Tamil Nadu India.
2Department of Botany, Government Arts College, Coimbatore, Tamil Nadu India.
3BI Biotech, Bengaluru-560079 India.Torrent Pharmaceuticals Ltd., Indore, MP India.
Corresponding author email: krishnavignesh.l@gmail.com
Article Publishing History
Received: 10/10/2024
Accepted After Revision: 25/12/2024
Antibiotic resistance is becoming a major threat and is increasing worldwide. The increasing prevalence of infections caused by antibiotic-resistant bacteria make the empirical treatment of Urinary Tract Infection (UTI) more difficult. The aim of the study is to find the antimicrobial susceptibility pattern of pathogens causing urinary tract infection and to help improve the effectiveness of empirical treatment. The pathogenic bacteria cultures were isolated from the patients with urinary tract infections in the district of Coimbatore, Tamil Nadu, India and its antimicrobial susceptibility pattern had been examined using ten different standard antibiotic discs. The antimicrobial activity was performed using Kirby Bauer disc diffusion method. The results had shown a considerable increase in the resistance pattern of the uropathogens.
Around 80% of the pathogens were being highly resistant to most of the antibiotics studied. Among the antibiotics, Imipenem being most effective followed by Gentamycin. Ampicillin was found to be the least effective. Most of the pathogens are resistant to more than three different classes of antibiotics. Present data reveal a steady emergence of multidrug resistance among the uropathogens in this region. Frequent multi-centre studies are required to understand the development of antimicrobial resistance of the pathogens as well as to know the effectiveness of the antibiotics prescribed. Situation urges to develop better antimicrobial agents with minimal or no side effects.
Antibiotic resistance, Antibiotics, Disc diffusion method, Urinary Tract Infections, Uropathogens
Lakshmanan K, Arumugam M, Mani R, Madhusoodnan M, Deepa O. Prevalence of Antimicrobial Resistance Amongst Uropathogens from Coimbatore: A Single-Centre Study. Biosc.Biotech.Res.Comm. 2024;17(4).
Lakshmanan K, Arumugam M, Mani R, Madhusoodnan M, Deepa O. Prevalence of Antimicrobial Resistance Amongst Uropathogens from Coimbatore: A Single-Centre Study. Biosc.Biotech.Res.Comm. 2024;17(4). Available from: <a href=”https://shorturl.at/Gj8c7“>https://shorturl.at/Gj8c7</a>
INTRODUCTION
Microbial resistance to drugs has become a global public-health threat compromising the efficacy of antimicrobial chemotherapy. The magnitude of this problem has recently been acknowledged by the World Health Organization, which has launched a global strategy for the containment of antimicrobial resistance. The emergence and spread of resistance are universally acknowledged to be associated with heavy consumption of antimicrobial agents in clinical and veterinary practices, and the prudent use of antibiotics has been considered to be mandatory for the preservation of their therapeutic effectiveness for as long as possible. Within this perspective, a combination of misuse and overuse of antimicrobial agents, along with overcrowding and poor sanitation, is among the reasons given to explain the exceedingly high resistance rates observed in low-income countries, (Okeke et al 1999, WHO 2001, Bartoloni et al 2004, Gajdács and Urban 2019, Kulkarni et al 2019, Anthony 2024 and PACCARB Report 2024).
The increasing prevalence of antimicrobial-resistant organisms are an important public health problem and is of particular concern for hospitals and other health care settings, (Fridkin et al 2002) Patterns of resistance in health care facilities are not uniform; even institutions in the same city may have quite different observed patterns of resistance for a given organism, (Lubowski et al 2001). In the United States, urinary tract infections result in about 8 million health practitioner visits per year. A 2013 record from the Centers for Disease Control and Prevention (CDC) revealed that greater than 2 million people within the US alone become ill each year as a result of antibiotic-resistant infections, and 23,000 die from such infections (Whiteman, 2014, Anthony 2024).
Resistance to antimicrobial agents among clinically important pathogens within the community and hospital settings has compromised therapy and requires constant monitoring of emerging patterns, (Jones 2003). The discovery of potent and safe antimicrobial agents is arguably the single greatest health care advance in history. The availability of these agents will rapidly reduce the morbidity and mortality associated with a host of formerly fatal diseases, (Rice 2008, PACCARB 2024).
Urinary tract infections are the most common site of microbial infections in human beings; it is estimated that 150 million UTI’S occur worldwide, resulting in more than 6 billion dollars in direct health care costs, (Stamm and Norrby 2001). Studies from different parts of India have indicated that UTI during pregnancy leads to low birth weight babies, increased perinatal mortality, and premature births along with many acute and chronic sequelae in the mother (Roy et al 1974). Recurrent urinary tract infection (rUTI) is a serious clinical problem, yet effective therapeutic options are limited, especially against multidrug-resistant uro-pathogens (O’Brien et al 2015, Anthony 2024).
The unrelenting increase in the prevalence of antimicrobial resistance is of great concern. The urgency of the problem is compounded by the recognition that fewer new antimicrobial agents are introduced each year, (Lautenbach 2009). The present study is designed to study the antimicrobial sensitivity pattern of different pathogens causing urinary tract infections among the people in Coimbatore.
MATERIAL AND METHODS
Collection and maintenance of pathogens: 50 various isolates of urinary tract infection causing pathogenic bacteria were collected from Sri Ramakrishna Hospital, Coimbatore from July to September 2013 and maintained on nutrient agar slants in cold room at 4°C.
Culture Media: Muller-Hinton Agar was prepared according to the manufacturer’s instruction, autoclaved, and dispensed at 20ml per plate in 12 X 12 cm Petri dishes. Set plates were incubated overnight to ensure sterility before use, (Lakshmanan et al 2012).
Antibacterial assay: Using the Agar disc diffusion method, Muller-Hinton agar plates were swabbed with the overnight grown culture of each microorganism. The antibiotic susceptibility discs were placed on each plate and incubated at 37°C for 24 hours (Lakshmanan 2013). The standard antibiotic discs used are Ampicillin (A), Cefpodoxime (CEP), Cefuroxime (CU), Cefepime (CPM), Piperacillin / Tazobactam (PT), Erythromycin (E), Gentamicin (G), Ciprofloxacin (CF), Co-Trimoxazole (CO) and Imipenem (I) were obtained from Hi-media. The Antibacterial activity was determined by measuring the diameter of zones of inhibition (mm) produced after incubation Krishnavignesh (2013). The susceptibility profiles of each pathogen against standard antibiotics were interpreted as per Clinical and Laboratories standard Institute (CLSI) guidelines.
Statistical Analysis: The results are expressed as the mean ± S.E.M. The results obtained from the present study were analyzed by using GraphPad Prism version 5.00 for Windows, GraphPad Software, San Diego California USA, www.graphpad.com.
RESULTS AND DISCUSSION
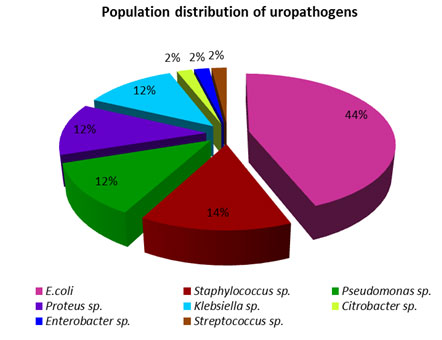
The collected bacterial population is comprised of 8 different species which are E. coli, Pseudomonas sp., Klebsiella sp., Proteus sp., Streptococcus sp., Staphylococcus sp., Citrobacter sp., Enterobacter sp. The population distribution displayed a clear dominance of E. coli over both the gram-positive and gram-negative uropathogens studied (Figure 1). The antimicrobial susceptibility pattern of each uropathogens was examined and their percentage was represented in charts (Figure. 1 – 8). In comparison with all the antibiotics investigated Imipenem is highly effective while Ampicillin being less effective.
Figure 1: Population distribution of uropathogens from samples of the study.
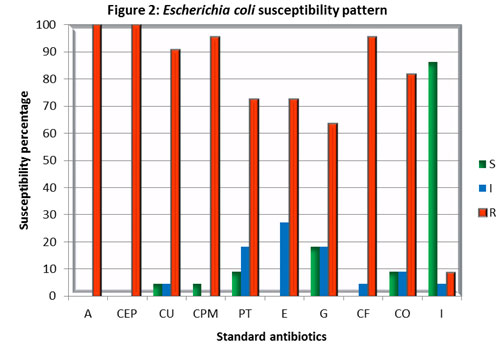
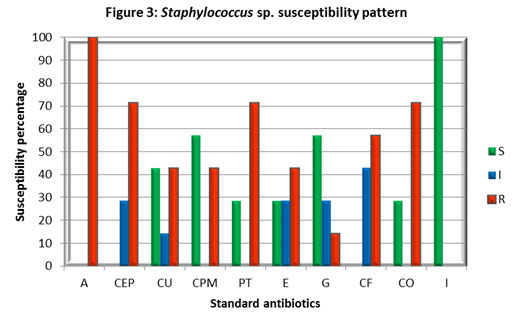
Foot note : S – Sensitive, I – Intermediate, R – Resistant, Ampicillin (A), Cefpodoxime (CEP), Cefuroxime (CU), Cefepime (CPM), Piperacillin / Tazobactam (PT), Erythromycin (E), Gentamicin (G), Ciprofloxacin (CF), Co-Trimoxazole (CO) and Imipenem (I).
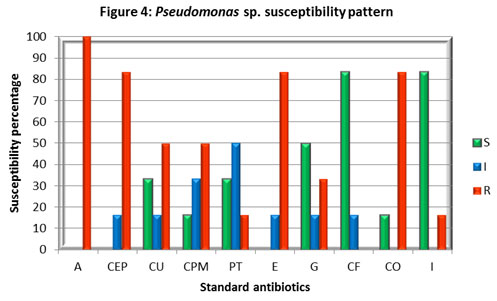
Foot note : S – Sensitive, I – Intermediate, R – Resistant, Ampicillin (A), Cefpodoxime (CEP), Cefuroxime (CU), Cefepime (CPM), Piperacillin / Tazobactam (PT), Erythromycin (E), Gentamicin (G), Ciprofloxacin (CF), Co-Trimoxazole (CO) and Imipenem (I).
Foot note : S – Sensitive, I – Intermediate, R – Resistant, Ampicillin (A), Cefpodoxime (CEP), Cefuroxime (CU), Cefepime (CPM), Piperacillin / Tazobactam (PT), Erythromycin (E), Gentamicin (G), Ciprofloxacin (CF), Co-Trimoxazole (CO) and Imipenem (I).
Foot note : S – Sensitive, I – Intermediate, R – Resistant, Ampicillin (A), Cefpodoxime (CEP), Cefuroxime (CU), Cefepime (CPM), Piperacillin / Tazobactam (PT), Erythromycin (E), Gentamicin (G), Ciprofloxacin (CF), Co-Trimoxazole (CO) and Imipenem (I).
Along with urinary tract infections, the resistance level of uropathogens posing a huge threat to the medical well-being of humans across the globe. Urinary tract pathogens in Madagascar are becoming increasingly resistant to commonly used antibiotics that are readily available at a low price. This poses a real problem for the treatment of community-acquired urinary tract infections (UTIs) in Madagascar (Randrianirina et al 2006).
Even India also experiences the same problem, Meta-analyses of the drug susceptibility results of various laboratories in India reveal an increasing trend in the development of resistance to commonly used antimicrobial agents in pathogens like Salmonella, Shigella, Vibrio cholerae, Staphylococcus aureus, Neisseria gonorrhoeae, N. meningitidis, Klebsiella, Mycobacterium tuberculosis, HIV, Plasmodium and others. One of the major concern is that there are no national data based on antimicrobial resistance in different pathogens yet known that the infectious disease burden in India is among the highest in the world, (WHO 2010, Kumar et al 2013).
In the present investigation, the population of uropathogens was comprised of Escherichia coli (44%) followed by Staphylococcus sp., (14%), Pseudomonas sp., (12%), Proteus species, (12%), Klebsiella species (12%), and 2% of Citrobacter species, Enterobacter species and Streptococcus sp. (Figure 1). It has shown that E. coli was to be the major causative of urinary tract infections. The same pattern of distribution of uropathogens was reported in another 5-year study of Gupta et al (1999) which showed the presence of Escherichia coli (86%) followed by Staphylococcus saprophyticus (4%), Proteus species (3%), Klebsiella species (3%), Enterobacter species (1.4%), Citrobacter species (0.8%), and Enterococcus species (0.5%). Other, less frequent isolates in aggregate caused 1.3% of infections. It indicates that E. coli is being predominant amongst the other uropathogens. E. coli cultures shown a high level of resistance towards Ampicillin (A), Ciprofloxacin (CF), Erythromycin (E), and Cefpodoxime (CEP) while sensitive to Imipenem (I) followed by Gentamycin (G) (Figure. 1).
These results were supported by several other studies that stated the majority of E. coli isolates resistant to ampicillin, trimethoprim-sulfamethoxazole, or ciprofloxacin, (Zhannel et al 2000). The prevalence of fluoroquinolone-resistant isolates of E. coli has been reported to be increasing over time in the United States and Canada. (Hooton 2003). 7.1% of urinary isolates of E. coli (2,763 of 38,835) were found resistant to three or more agents and considered multidrug-resistant. Among the multidrug-resistant isolates, 97.8% were resistant to ampicillin, 92.8% were resistant to trimethoprim-sulfamethoxazole, 86.6% were resistant to cephalothin, 38.8% were resistant to ciprofloxacin, and 7.7% were resistant to nitrofurantoin, ( Karlowsky et al 2002, Sahm et al 2001).
It shows that E. coli is gaining resistance to multiple antibiotics. The results of this study also raises the alarm of the emergence of multidrug resistance among the E. coli strains in Coimbatore. This may be due to the increase in strains harbouring an extended-spectrum β lactamase. Staphylococcus sp. cultures showed higher resistance towards Ampicillin (A), Piperacillin/Tazobactam (PT), Co-trimoxazole (CO), Cefpodoxime (CEP), while Imipenem (I) found to be highly effective with 100% inhibition followed by Gentamicin (G) and Cefepime (CPM). The study indicates the emergence of multidrug resistance among Staphylococcus cultures isolated from patients with urinary tract infections.
In support of the current findings already a high level of MRSA was reported in our country, which found 41% of methicillin resistance among 26310 isolates (India et al 2013). In India, Gram-positive infections, particularly methicillin-resistant Staphylococcus aureus (MRSA) prevalence among invasive S. aureus isolates, have been increased exponentially from 29% in 2009 to 47% in 2014. Apart from MRSA, the rising prevalence of vancomycin-resistant enterococci (VRE), which ranges from 1 to 9% in India, has raised concerns, (Kulkarni et al 2019).
The majority of the Pseudomonas cultures investigated in the study were highly sensitive to Imipenem (I), and Ciprofloxacin (CF), followed by Gentamicin (G). While 50% of the isolates found intermediate to Piperacillin / Tazobactam (PT) and 33% were sensitive and found resistant to other antibiotics studied. This result was correlated with the study conducted in Pakistan, (Shah et al 2015) which reported 10% resistance towards Imipenam, 19.6% towards Piperacillin / Tazobactam (PT), 35% vs Gentamicin (G), and varies with the findings against 99% vs CU; 50% vs CF whereas we observed 50% and 100% resistance against CU and CF respectively.
This variation may be due to the indiscriminate use of antibiotics in the respective geographical areas. Besides the few contradictions, all these studies substantiate the development of multidrug resistance Pseudomonas strains. Infection caused by this organism is difficult to treat because of the presence of its innate resistance to many antibiotics (β-lactam and carbapenem group of antibiotics), and its ability to acquire further resistance mechanisms to multiple classes of antibiotics, including Beta-lactams, aminoglycosides, and fluoroquinolones, (Pachori et al 2019).
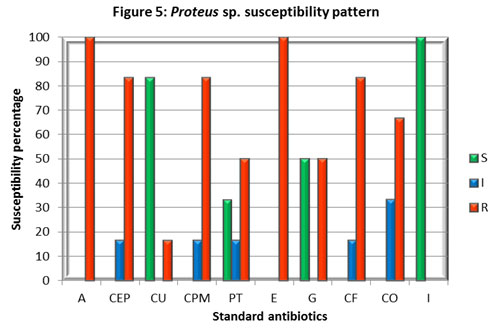
Proteus isolates were shown higher sensitivity to Imipenem (I), Cefuroxime (CU) followed by 33% of sensitivity towards Piperacillin / Tazobactam (PT), and it has been found resistant to all other antibiotics investigated. The emergence of multidrug resistance observed among Proteus sp. in this study in Coimbatore district contradicts the findings of multicenter surveillance conducted in Taiwan that revealed decreased susceptibility of P. mirabilis to some broad-spectrum antibiotic particularly to the resistance rate against Imipenem. Besides the current study found zero percent resistance compared to 51.8%, however it correlates with the majority of the findings related to the increasing resistance level reported against different antibiotics including 3rd-generation cephalosporins and ciprofloxacin, in the past decade. Similar results were reported in Banglore, India found more than 80% of resistance in Proteus sp. This finding indicates the development of antibiotic resistance among Proteus sp. against multiple antibiotics. This might be due to a higher prevalence of ESBL- and/or AmpC β-lactamase- producers, (Wang et al, 2014, Navneeth et al 2002).
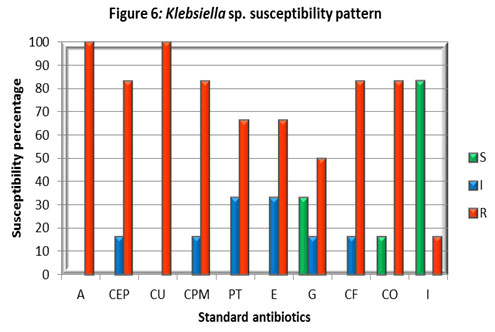
Klebsiella isolates showed good susceptibility to Imipenem (I) and moderately susceptible to Gentamicin (G) while it has shown resistance to all other antibiotics studied. A report in Uganda found concurrence with the current study showing 82% of Klebsiella sp. was resistant to at least three classes of antibiotics, (Stanley et al 2018).
In Tanzania, the resistance level of Klebsiella spp strains was cotrimoxazole (92.6%), ciprofloxacin (25.0%), and gentamicin (38.2%) but our study in Coimbatore district displayed a considerably higher level of resistance of 83.3%, 83.3%, and 50% against the respective antibiotics. This might be due to the production of ESBL among the strains, (Moyo et al 2020).
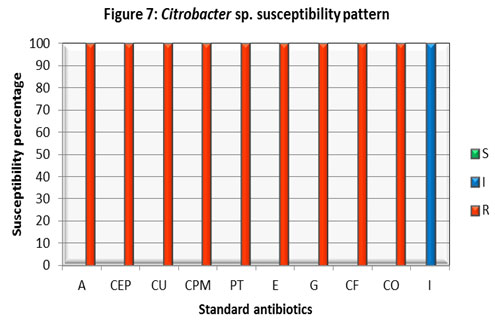
All the above studies found Imipenem is being effective against Klebsiella sp. Citrobacter isolate found intermediate only to Imipenem (I) and exhibited resistance to all other antibiotics tested. The current study was found parallel with another which reported the majority of the urinary tract Citrobacter isolates were found to be resistant to cefotaxime, cephalexin, norfloxacin, ciprofloxacin, and the aminoglycosides. This may be because of the widespread use of broad-spectrum antibiotics, leading to selective survival advantage of pathogens, (Metri et al 2013).
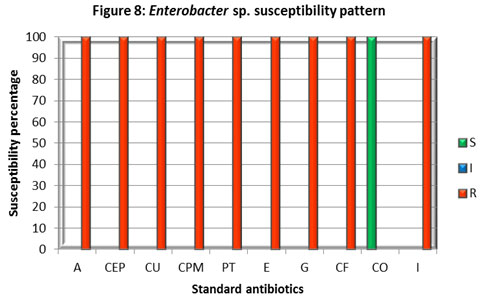
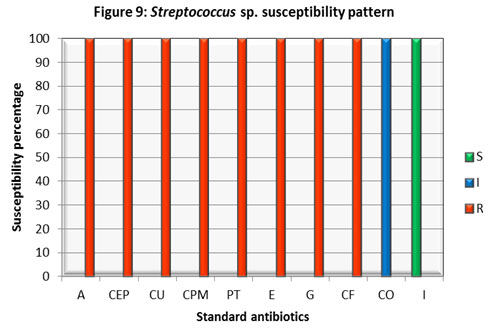
Enterobacter isolate was susceptible to Co-trimoxazole (CO) while it was found resistant to all other antibiotics inspected. The most crucial finding was its resistance to Imipenem. Streptococcus isolate found sensitive to Imipenem (I) and intermediate to Co-trimoxazole (CO), to the rest of antibiotics, it displayed resistance. Citrobacter., Enterobacter, and Streptococcus isolates displayed a higher rate of resistance against different classes of antibiotics. A study in Hungary also reported the emergence and spread of ESBL-and/or carbapenemase-producing Enterobacter (Gajdács et al 2019).
Our study has shown Imipenem (I) as an effective antibiotic against the majority of the uropathogens investigated followed by Gentamycin (G). Though most of the organisms were not sensitive to Ciprofloxacin (CF) it was found intermediate in a considerable volume of the microbial population investigated. Piperacillin / Tazobactam (PT), Cefuroxime (CU), and Cefepime (CPM) had shown moderate activity. Co-Trimoxazole (CO), Erythromycin (E) and Cefpodoxime (CEP) were found to be least effective. Most of the reports worldwide found Imipenem to be one of the most effective antibiotics. Ampicillin resistance pattern varies among countries, was reported to be Ethiopia (17%), Italy (36%), UK (23%), USA (43%), Canada (33%), Europe (50%) and Norway (25%), (Bitiv et al 2017).
In contrast to the above findings our study in Coimbatore district, Tamil Nadu, India was found to be 100% against both the gram-negative as well as gram-positive uropathogens, these findings were supported by another study conducted at Banglore, India reported a 93% resistance among the gram-negative uropathogens, (Navneeth et al 2002). Studies from African countries and Iran also reported 100% and 89.29% resistance respectively against ampicillin, (GLASS Report 2017).
Though Imipenem was found to be an effective antibiotic amongst most of the organisms, still this study found a small fraction of microbial population resistant to it. It is an alarming signal since carbapenemase-producing pathogens are a significant clinical and public health concern that will drastically limit the therapeutic armamentarium, (Gajdacs and Urban 2019).
Several factors contribute to the emergence of antimicrobial resistance, they are indiscriminate use of antibiotics, intrinsic antibiotic resistance like efflux systems and producing antibiotic-inactivating enzymes, acquired antibiotic resistance like mutations, adaptive antibiotic resistance like biofilm mediated resistance, and microbial genomic plasticity. The constant changing of the microbial genome is a bigger threat than the infection itself. There are several mechanism microbes utilize to alter their genomes such as horizontal gene transfer, transposon-mediated genomic alterations and promiscuous nature of the gene and genomic products.
Some of the world’s most common and potentially most dangerous infections are proving drug-resistant and most worrying of all is pathogens don’t respect national borders. That’s why WHO is encouraging all countries to set up good surveillance systems for detecting drug resistance that can provide data to this global system, (GLASS Report 2017 WHO 2018). This emphasizes the importance of the local resistance data which is crucial to any part of the world. Though the sample size of this study is small it provides the much needed local resistance data which reveals the first details on population distribution among the uropathogens and its antimicrobial resistance pattern towards the different class of antibiotics in the Coimbatore district, Tamil Nadu. With these initial insights, it warrants a detailed multicentric study on the surveillance of antimicrobial resistance patterns of uropathogens to design an effective protocol.
CONCLUSION
The present study revealed that the uropathogens are steadily gaining resistance towards most of the commonly used antibiotics. This is a clear warning signal over the emergence of multidrug resistance among the uropathogens in Coimbatore, India as well as all across the globe. Concurrent resistance to antimicrobials of different structural classes has arisen in a multitude of bacterial species and complicates the therapeutic management of urinary tract infections. Frequent prevalence studies on pathogens causing urinary tract infections are warranted to understand the ever-changing antimicrobial trends in their respective regions. Growing multidrug resistance limits the therapeutic options. This scenario urges the scientific and medical community to develop additional effective antibiotics in order to serve the community in a better way. With all this, we are in a situation to frame a strict operational policy of antimicrobial therapies and standard treatment guidelines.
Funding: Nil
Conflict of interest statement: Authors declare no conflict of interest.
Data Availability: All data are available with the corresponding author
REFRENCES
Anthony D. (2024) Catalyzing Policy Action to Address Antimicrobial Resistance: Next Steps for Global Governance South Centre Geneva 19 Switzerland
Bartoloni A, Bartalesi F, Mantella A, Dell’Amico E, Roselli M, Strohmeyer M, Barahona HG, Barrón VP, Paradisi F, Rossolini GM. (2004) High prevalence of acquired antimicrobial resistance unrelated to heavy antimicrobial consumption. Journal of Infectious Diseases. 2004;189(7):1291-4.https://doi.org/10.1086/382191.
Bitew A, Molalign T, Chanie M.(2017) Species distribution and antibiotic susceptibility profile of bacterial uropathogens among patients complaining urinary tract infections. BMC infectious diseases. 2017;17(1):654.
Fridkin SK, Hill HA, Volkova NV, Edwards JR, Lawton RM, Gaynes RP, McGowan Jr JE. (2002) Temporal changes in prevalence of antimicrobial resistance in 23 US hospitals. Emerging infectious diseases. 2002;8(7):697.
Gajdács M, Urbán E. (2019) Resistance Trends and Epidemiology of Citrobacter-Enterobacter-Serratia in Urinary Tract Infections of Inpatients and Outpatients (RECESUTI): A 10-Year Survey. Medicina. 2019;55(6):285.
Global antimicrobial resistance surveillance system (GLASS) report: early implementation 2016-2017. Geneva: World Health Organization; 2017. Licence: CC BY-NC-SA 3.0 IGO. https://www.who.int/mediacentre/news/releases/2018/antibiotic-resistance-found/en/.
Gupta K, Hooton TM, Wobbe CL, Stamm WE (1999) The prevalence of antimicrobial resistance among uropathogens causing acute uncomplicated cystitis in young women. International journal of antimicrobial agents. 1999;11(3-4):305-8.
Hooton TM. Fluoroquinolones and resistance in the treatment of uncomplicated urinary tract infection. International journal of antimicrobial agents. 2003;22:65-72.
India SJ, Ray P, Manchanda V, Bajaj J, Chitnis DS, Gautam V, Goswami P, Gupta V, Harish BN, Kagal A, Kapil A. Methicillin resistant Staphylococcus aureus (MRSA) in India: prevalence & susceptibility pattern. The Indian journal of medical research. 2013;137(2):363.
Jones RN (2003) Global epidemiology of antimicrobial resistance among community-acquired and nosocomial pathogens: a five-year summary from the SENTRY Antimicrobial Surveillance Program (1997-2001). In Seminars in respiratory and critical care medicine 2003 (Vol. 24, No. 01, pp. 121-134). Thieme Medical Publishers, New York USA.
Karlowsky JA, Kelly LJ, Thornsberry C, Jones ME, Sahm DF. Trends in antimicrobial resistance among urinary tract infection isolates of Escherichia coli from female outpatients in the United States. Antimicrobial agents and chemotherapy. 2002;46(8):2540-5.
Krishnavignesh L, Mahalakshmipriya A, Ramesh M. In vitro analysis of phytochemical screening and antimicrobial activity of Parthenium hysterophorus L. against pathogenic microorganisms. Asian J Pharm Clin Res. 2013;6(5):41-4.
Kulkarni AP, Nagvekar VC, Veeraraghavan B, Warrier AR, TS D, Ahdal J, Jain R. Current Perspectives on Treatment of Gram-Positive Infections in India: What Is the Way Forward?. Interdisciplinary perspectives on infectious diseases. 2019;2019.
Kumar SG, Adithan C, Harish BN, Sujatha S, Roy G, Malini A. Antimicrobial resistance in India: A review. Journal of natural science, biology, and medicine. 2013;4(2):286.
Lakshmanan K, Arumugam M, Madhusudanan M. Evaluation of anti microbial activity and phytochemical screening of Tridax procumbens and Aerva lanata (L.) Juss. ex Schult against uropathogens. J Pharm Res. 2012;5:2377-9.
Lakshmanan K, Arumugam M, Mani R. Phytochemical Screening and In Vitro Antimicrobial activity of Vitex negundo L. var. purpurascens Sivar. and Mold. against pathogenic microorganisms. Drug Invention Today. 2012;4(12):667-70.
Lautenbach E. Antimicrobial resistance in gram-negative pathogens: crafting the tools necessary to navigate the long ascent out of the abyss. The Journal of infectious diseases. 2009 Sep 1;200(6):838-40..
Lubowski TJ, Woon JL, Hogan P, Hwang CC. Differences in antimicrobial susceptibility among hospitals in an integrated health system. Infect Control Hosp Epidemiol. 2001; 22:379-82.
Metri BC, Jyothi P, Peerapur BV. Antibiotic resistance in Citrobacter spp. isolated from urinary tract infection. Urology annals. 2013;5(4):312.
Moyo SJ, Aboud S, Kasubi M, Lyamuya EF, Maselle SY. Antimicrobial resistance among producers and non-producers of extended spectrum beta-lactamases in urinary isolates at a tertiary Hospital in Tanzania. BMC research notes. 2020 ;3(1):348.
Navaneeth BV, Belwadi S, Suganthi N. (2002) Urinary pathogens’ resistance to common antibiotics: a retrospective analysis. Tropical doctor. 2002;32(1):20-2.
O’Brien VP, Hannan TJ, Schaeffer AJ, Hultgren SJ. (2015) Are you experienced? Understanding bladder innate immunity in the context of recurrent urinary tract infection. Current opinion in infectious diseases. 2015;28(1):97.
Okeke IN, Lamikanra A, Edelman R. Socioeconomic and behavioral factors leading to acquired bacterial resistance to antibiotics in developing countries. Emerging infectious diseases. 1999;5(1):18.
PACCARB Report (2024) A United Front: Collaborative Global Leadership To Combat Antimicrobial Resistance A Report With Recommendations May 2024
Pachori P, Gothalwal R, Gandhi P. Emergence of antibiotic resistance Pseudomonas aeruginosa in intensive care unit; a critical review. Genes & diseases. 2019;6(2):109.
Randrianirina F, Soares JL, Carod JF, Ratsima E, Thonnier V, Combe P, Grosjean P, Talarmin A. Antimicrobial resistance among uropathogens that cause community-acquired urinary tract infections in Antananarivo, Madagascar. Journal of Antimicrobial Chemotherapy. 2006;59(2):309-12.
Rice LB. Federal funding for the study of antimicrobial resistance in nosocomial pathogens: J Infect Disease 2008 Apr 15;197(8):1079-81. doi: 10.1086/533452. DOI: 10.1086/533452
Roy, SK, Sinha, GR, and Quadros, M. The study of bacteriuria in pregnancy. J. Obst. Gynec. Ind. 1974; 245-251.
Sahm DF, Thornsberry C, Mayfield DC, Jones ME, Karlowsky JA. Multidrug-Resistant Urinary Tract Isolates of Escherichia coli: Prevalence and Patient Demographics in the United States in 2000. Antimicrobial agents and chemotherapy. 2001;45(5):1402-6.
Shah DA, Wasim S, Abdullah FE. Antibiotic resistance pattern of Pseudomonas aeruginosa isolated from urine samples of Urinary Tract Infections patients in Karachi, Pakistan. Pakistan journal of medical sciences. 2015;31(2):341.
Stamm WE, Norrby SR. Urinary tract infections: disease panorama and challenges. The Journal of infectious diseases. 2001;183(Supplement_1):S1-4.
Stanley IJ, Kajumbula H, Bazira J, Kansiime C, Rwego IB, Asiimwe BB. Multidrug resistance among Escherichia coli and Klebsiella pneumoniae carried in the gut of out-patients from pastoralist communities of Kasese district, Uganda. PloS one. 2018;13(7):e0200093.
Wang JT, Chen PC, Chang SC, Shiau YR, Wang HY, Lai JF, Huang IW, Tan MC, Lauderdale TL. Antimicrobial susceptibilities of Proteus mirabilis: a longitudinal nationwide study from the Taiwan surveillance of antimicrobial resistance (TSAR) program. BMC infectious diseases. 2014;14(1):486.
Whiteman H. Antibiotic resistance: how has it become a global threat to public health. Medical News Today. 2014;10.
World Health Organization. High levels of antibiotic resistance found worldwide, new4 data shows. Saudi Medical Journal. 2018;39(4):430-1.
World Health Organization. Prevention and containment of antimicrobial resistance. Report of a regional meeting Chiang Mai, Thailand, 8th to 11th of June 2010. Available from http://www.searo.who.int/LinkFiles/BCT_Reports_SEA-HLM-408.pdf .
World Health Organization. WHO global strategy for containment of antimicrobial resistance. Geneva: World Health Organization; 2001.
Zhanel GG, Karlowsky JA, Harding GK, Carrie A, Mazzulli T, Low DE, Hoban DJ. A Canadian national surveillance study of urinary tract isolates from outpatients: comparison of the activities of trimethoprim-sulfamethoxazole, ampicillin, mecillinam, nitrofurantoin, and ciprofloxacin. Antimicrobial agents and chemotherapy. 2000;44(4):1089-92.


