Department of Biotechnology,College of Agriculture and Natural Resource,
Debre Berhan University, P. O. BOX 445 Debre Berhan, Ethiopia
Corresponding author email: fkdms2008amu@gmail.com
Article Publishing History
Received: 27/10/2023
Accepted After Revision: 15/12/2023
Wheat (Triticum aestivum L) is the main source of nutrition and feeds more than 30% of the world’s population. It is grown in various environments, providing most humans with around 20% of their calories and protein. Genome editing holds countless promise to accelerate the development of improved crop varieties including wheat via providing powerful tools to modify the genomic regions that controlling major agronomic traits. CRISPR/Cas9 system is an effective method for targeted genome editing. CRISPR/Cas9 allows researchers to target multiple homoeoalleles simultaneously and it enables the production of targeted mutations in all copies of a gene.
The CRISPR-Cas and associated technologies derived from the naturally occurring prokaryotic CRISPR immune system and it has been used to target multiple homoeoalleles simultaneously and accelerate progress in functional genomics and molecular breeding in wheat. CRISPR/Cas9 is widely used to improve agricultural traits by knocking out unwanted genes or genes conferring undesirable phenotypes. Targeted knockout wheat with foreign DNA is generated in the T0 generation. Ultimately, the foreign DNA can be segregated by selfing and crossing. A targeted gene-modified plantlet without foreign DNA is generated in the T0 generation.
This approach has been reported in wheat for the first time. DNA free genome-edited wheat plants have been generated. Genes required for genic male sterility are identified, CRISPR/Cas9- mediated disruption of these genes will enable the rapid production of male-sterile wheat. This represents a promising method for manipulating recessive sterility genes to capture heterosis in wheat. CRISPR-based genome editing will bring functional genomics and rational design-based molecular breeding of polyploid wheat to the forefront of wheat biology. Transgene-free, gene edited wheat will play a critical role in addressing environmental issues while promoting sustainable agriculture.
CRISPR/Cas9, DNA, Editing, Genome, Improvement, Wheat
Namo F.M. Optimizing Genome Editing for Wheat Genetic Improvement: Current Status and Future Prospective. Biosc.Biotech.Res.Comm. 2023;16(4).
Namo F.M. Optimizing Genome Editing for Wheat Genetic Improvement: Current Status and Future Prospective. Biosc.Biotech.Res.Comm. 2023;16(4). Available from: <a href=”http://surl.li/pefvi“>http://surl.li/pefvi</a>
INTRODUCTION
Wheat (Triticum aestivum L) is the main source of nutrition and feeds more than 30% of the world’s population (Wang et al., 2019). The demand for wheat with high-quality traits has increased globally due to the growing population and the rising living standards in countries worldwide (Kumar et al., 2019). The presence of wheat gluten gives the dough viscoelasticity and ductility, and it can be processed into a variety of foods to meet people’s needs (Veraverbeke and Delcour, 2002). Common wheat is a keystone crop species. It is grown in various environments, providing most humans with around 20% of their calories and protein (Uauy et al. 2017); thus, it occupies an important position in food security. As the global population increases, improving the yield of wheat is critical to ensure future availability. Geneticists have exploited natural or artificial wheat variations for breeding. Indeed, conventional breeding approaches have played a major role in increasing grain yields and quality based on broad genetic variations in wheat (Nadolska-Orczyk et al. 2017).
However, wheat is an allohexaploid (2n = 6 9 = 42, AABBDD); it harbors three closely related sub genomes inherited from three homoeologous ancestors (Petersen et al. 2006). Thus, most wheat genes have three similar but not identical copies, with functional redundancy and complementarity among the A, B, and D genomes. As a result, the probability of the simultaneous mutation of genes in the A, B, and D genomes by natural processes or induced mutagenesis is very low. Therefore, the complex polyploid nature of wheat has hindered the development of functional genomics and breeding, especially compared to other cereals, such as rice and maize.
Genome editing holds countless promise to accelerate the development of improved crop varieties via providing powerful tools to modify the genomic regions controlling major agronomic traits. The actual deployment of these technologies for crop improvement requires further optimization of the components of the genome editing systems and tools for different applications. The functionality of the various components of the genome editing tools like CRISPR/Cas9 system was authenticated using the wheat protoplast transformation assay followed by the next-generation sequencing (NGS) of the targeted genomic regions. The wheat codon optimized Cas9 was revealed to be effective tool for the targeted gene editing in the wheat genome.
The recent emergence of genome-editing technologies has transformed basic biological studies and practical biotechnological applications. Among the major genome-editing tools such as zinc finger nuclease (ZFN), transcription activator-like effector nuclease (TALEN), and clustered regularly interspaced short palindromic repeats (CRISPR)-CRISPR-associated nuclease (Cas), CRISPRCas- mediated genome-editing technique has become the method of choice for most of the laboratories due to its simplicity, affordability, and high success rate. Although many laboratories around the world have rapidly adopted conventional CRISPRCas9-mediated approaches for targeted mutation and gene knockout via the non-homologous end-joining (NHEJ) repair, precise genome editing with CRISPR-Cas9 remains challenging to achieve (Molla and Yang, 2019).
Typically, precise changes in the genome require the use of homology-directed repair (HDR), which involves the supply of an exogenous donor template with the desired changes. However, the efficiency of HDR-mediated genome editing is extremely low in higher plants due to the difficulties in delivering adequate repair templates and the low rate of homologous recombination. Moreover, HDR occurs only during S and G2 phase of the cell cycle.CRISPR/Cas9 system is an effective method for targeted genome editing, and its efficiency has been shown in several plant species (Endo et al., 2015).
Although this system is relatively easy to use and more precise compared to other genome editing technologies, there are still some issues, particularly for polyploidy plants, such as editing efficiency and off-target mutation rate. Here, a series of experiments were conducted to show the efficient genome editing with CRISPR/Cas9 system in wheat protoplast and the results confirmed that CRISPR/Cas9 system is a promising tool for further targeted editing of wheat genome (Jun et al., 2021).
The CRISPR-Cas9-mediated genome-editing technology provides unprecedented tools to precisely edit DNA sequences in animals and plants (Gao, 2018, Mali et al., 2013). This technology requires expression of the Cas9 protein, production of a guide RNA (gRNA) that complements the target DNA sequences, and the existence of an NGG protospacer adjacent motif (PAM) site in the target sequence (Cong et al., 2013, Mali et al., 2013). Briefly, genome editing mediated by CRISPR-Cas9 utilizes a 20-bp gRNA that directs the Cas9 nuclease to the target site by base pairing.
Cas9 cuts the target site to generate a double-strand break (DSB). Mutations are introduced during the DNA repairing process. Because of its simplicity, CRISPR-Cas9 has been widely adopted. Several CRISPR vectors have been developed for genome editing in plants (Ding et al., 2018). It was proven that CRISPR-Cas9-mediated genome editing technology could successfully generate various heritable mutations in plant species (Bortesi et al., 2016).
After transformation, the first mission is to isolate primary transformants with expected mutations. To this end, restriction enzyme digestion or sequencing of the PCR amplicons is typically performed (Bortesi et al., 2016). However, both methods are time- and money consuming and laborious. Next, mutants without the T-DNA insertion may need to be identified, which are favored in both basic and applied researches because of the following reasons. First, prolonged existence of CRISPR-Cas9 in the mutants would greatly increase the risk of producing off-target mutations.
Second, transgene-free materials are more easily accepted by the public. Cas9-free mutants could be obtained by self-crossing or backcrossing. In B. napus, for instance, by using gene-specific primers (such as Cas9), 10.9% (58/530) of the mutant plants in the T1 generation lost the Cas9 transgene by self-crossing (Yang et al., 2017). a ratio lower than what is expected, which need larger populations to get Cas9-free mutant. Therefore, an easy method to screen for the mutants in need in both primary transformants and their offspring is highly required.
Application of the CRISPR/Cas9 system requires the DNA sequences of the target genes. Given the availability of the annotated wheat genome and the elucidation of a growing number of genes controlling important agronomic traits in other plants, it is easy to isolate orthologous genes in wheat based on homology based cloning. In addition, CRISPR/Cas9 allows researchers to target multiple homoeoalleles simultaneously and it enables the production of targeted mutations in all copies of a gene; thus, the system holds great promise in the characterization of genes endowing important agronomic traits in polyploid wheat. Furthermore, it has been used to modify multiple genes controlling different agronomic traits in wheat. This technology will bring a new dawn to wheat biology and breeding programs. In this review, we briefly outline the utilization of the CRISPR/Cas9 system, with an emphasis on the most important breakthroughs thus far.
CRISPR/Cas9 contains two major components: a sgRNA, which is responsible for recognizing target DNA, and the Cas9 endonuclease, which is responsible for generating DSB at predesigned target DNA site. Cas9 from Streptococcus pyogenes (SpCas9) was the first well-characterized RNA-guided endonuclease. It is a multifunctional protein that contains two nuclease domains: the HNH domain and RuvC-like domain. Each of them cuts one DNA strand, generating blunt-end DSBs; this triggers endogenous DNA repair systems, resulting in targeted mutants.
The only prerequisite for applying CRISPR/Cas9 to a given site is the presence of a protospacer-adjacent motif (PAM; NGG for SpCas9) next to the sequence of interest. For different target sites, Cas9 is constant; we can only change the guide sequence in the sgRNA. Crop improvement aims to increase crop yield and resistance to biotic and abiotic stress, as well as quality and nutritional value. Crop yield has been significantly increased through advanced agricultural technologies over several decades. Crop quality has been a greater concern of consumers since it is directly associated with human health by providing multiple nutrients such as proteins, fiber, vitamins, minerals, and bioactive compounds, (Lavin, and Lloyd, 2012).
Genome editing can create predictable and inheritable mutations in specific sites of genome, with the lowest probability of off-target and no integration of exogenous gene sequences. GE-mediated DNA modifications encompass deletions, insertions, single nucleotide substitution (SNPs), and large fragment substitution. Four site-directed nuclea (SDN) families are involved in a nucleotide excision mechanism: homing endonucleases 0or mega-nucleases (HEs) (Cohen et al., 1998), Zinc-Finger Nucleases (ZFNs) [Bibikova, et al., 2002), transcription activator like effector nucleases (TALENs) (Christian et al., 2010) and CRISPR-associated protein (CAS) (Cong et al., 2013).
The number of cases in crop improvement using GE has increased significantly. Among the various target traits for crop improvement, crop quality is one of the highest objectives. Here, we summarized the recent progress in CRISPR/Cas9-mediated crop quality improvement and provide further discussion on the future application of GE.
CRISPR/Cas9 Gene-Editing System in Plants: CRISPR/Cas systems have been divided into two classes and five types according to the classification of the CAS protein. The type II CRISPR/SpCas9 system from Streptococcus pyogenes has been modified and developed as versatile GE tools for different applications (Hsu et al., 2014). It consists of two core components: the guide RNA (gRNA or sgRNA) and the Cas9 protein. The gRNA constitutes CRISPR RNA (crRNA) and trans-activating crRNA (tracrRNA). The former contains a~20 nt fragment (also known as a spacer, complementary to a specific site of target genes), followed by a protospacer adjacent motif (PAM) in the target genes of interest. Under the guidance of gRNA, Cas9 nuclease creates DSBs at ~3 bp upstream of the PAM motif (Jinek, et al., 2012).
The cleavage repaired in NHEJ way, usually results in gene knockout or loss of protein function (Liu et al., 2019). Alternatively, when an exogenous DNA repair template is provided, HDR can be triggered, resulting in the introduction of the repair template into a target genomic region [9]. In plants, CRISPR/Cas9-based gene-editing consists of multiple steps including the selection of target sites, designing and synthesis of sgRNA, delivery of transformation carrier or ribonucleoprotein (RNP) in plant cells, transformation, and screening of gene-edited plants. At present, the plant CRISPR/Cas9 and its derived system have shown various genome-editing ability, such as gene knock-in, knockout, knockdown, and expression activation as well. In addition, simultaneous editing on multiple genes has contributed to pathway-level research.
The CRISPR-Cas and associated technologies derived from the naturally occurring prokaryotic CRISPR immune system are definitely revolutionary in studying basic biology and manipulating
genomes of diverse organisms. These robust, reproducible, and easy-to-use technologies allow the manipulation or modification of genome in several ways, including, but not limited to (a) by simply incorporating random mutation (insertion or deletion) through non-homologous end joining to disrupt gene(s); (b) by generating targeted point mutations in genes using precise base editors; and (c) by a whole gene insertion employing the cell’s homology directed repair pathway. Improvement in protocols, higher access to CRISPR-Cas tools, and necessary changes in the global regulatory environments are needed for the broader application of this frontier technology in diverse areas like production of energy, health, environment, medicine, and sustainable food production in changing climate (Tofazzal et al., 2020).
Figure 1: The workflow used in this study is workflow of generation CRISPR-Cas9
genome-editing transgenic plants, from target sgRNAs designing, plant
transformation, and isolating positive mutant plants
Figure 2: Design of the primer sets containing gene-specific sgRNAs. (a) At the “submit” page, select a
genome you are studying, and input gene locus, chromosome position, or DNA sequence in
FASTA format. Other parameters are set as default. A few seconds after submission, the
result is shown in web browser, which contains sgRNA sequence information,
including GC content, on-target score, off-target score, and restriction
endonuclease sites. (b) Choose the sgRNA sequences, and replace
19-nt N in the forward primers with 19-nt target sequencesin front
of PAM (NGG) and 19-nt N in the reverseprimers with reverse
complement sequences of 19-nt target sequences in front of PAM (NGG)

Figure 3: Construction of pKSE401G. (a) The CRISPR-Cas9 vector pKSE401G is modified
from pKSE401 (Xing et al., 2014). The 35S-sGFP-terminator cassette is amplified from
pK7GWIWG2D 30. (Karimi et al., 2007) using primers 35S-GFP-Ter-F and 35SGFP- Ter-R
and inserted into the PmeI site of pKSE401 by the Gibson Assembly method
(Gibson et al., 2009). (b) The map of pCBC-DT1T2 (Xing et al., 2014)
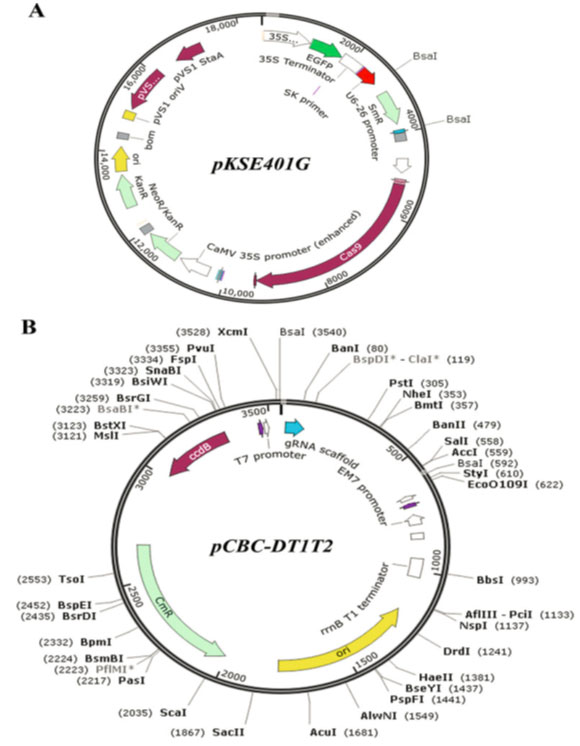
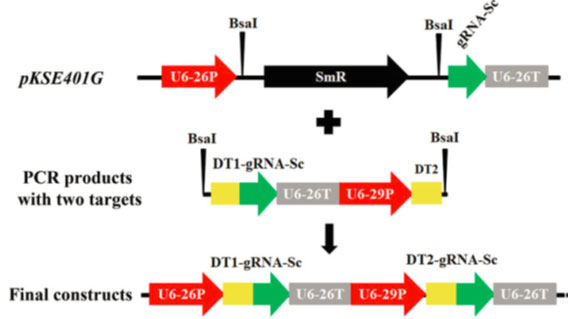
Figure 4: The gRNA modules used for the assembly of two gRNA expression cassettes. Examples
of the assembly of two-gRNA expression cassettes for dicots using the gRNA modules.
Using pCBC-DT1T2 as the template, two AtU6 promoter-sgRNA-AtU6 terminator
cassettes were amplified by PCR, and the PCR fragments were then inserted
into pKSE401G by Golden Gate Assembly. U6-29p and U6-26p are two
Arabidopsis U6 gene promoters; U6-29t and U6-26t, corresponding
Arabidopsis U6 gene terminators with downstream sequences,
respectively. gRNA-Sc, gRNA scaffold; DT1/2, dicot target-1/2
Analyzing genome editing events by next-generation sequencing (NGS): The transient expression in the transformed protoplasts provides a simple and rapid method for assessing the editing capability of the CAS9/gRNA constructs (Upadhyay et al., 2013, Shan et al., 2013). This approach has adopted to screen gRNAs for the ability to generate Cas9-mediated changes in the wheat genome. Further, the protoplast expression assay was combined with NGS for the rapid and cost-effective analysis of multiple genomic regions. This strategy was used to evaluate the gRNAs designed to target four genes controlling domestication (Q gene) (Simons et al., 2006), seed development (TaGW2) (Su et al., 2011) and disease resistance (TaLpx-1 and TaMLO) phenotypes in wheat (Wang et al., 2014, Nalam et al., 2015).
The editing specificity of the designed gRNAs was certified by peer review) is the author/funder. All rights reserved. No reuse allowed without permission. The A and D genome copies of the Q gene were successfully targeted by three gRNAs on the 5’-end, and only the A genome copy was targeted by two gRNAs on the 3’-end. Only one of the two target sites in TaGW2 was edited in all three wheat genomes. Both gRNAs designed for the TaLpx-1 gene produced deletions in the B and D genomes, whereas no editing events were detected for the target in the A genome due to its high divergence from the guide sequence. The gRNA targeting TaMLO induced mutations only in the wheat a genome, as previously reported (Wang et al., 2014).
Current Status of Genome Editing in Wheat: CRISPR/Cas9 has been used to target multiple homoeoalleles simultaneously and it will accelerate progress in functional genomics and molecular breeding in wheat.
Generating CRISPR-edited DNA-free wheat: CRISPR/Cas9 is widely used to improve agricultural traits by knocking out unwanted genes or genes conferring undesirable phenotypes. However, this process usually involves transgenic intermediates, which causes regulatory concerns and is not accepted worldwide (Zhang et al. 2020). For public acceptance, gene removal or bypassing foreign elements to edit endogenous genes is a good choice (He and Zhao, 2020). Based on the reagents needed for CRISPR-mediated editing, there are two main ways to produce CRISPR-edited DNA-free plants.
In the vector-based method, a vector is delivered into wheat callus using Agrobacterium or particle bombardment. It then integrates into the genome and the encoded genome editing elements are expressed, enabling targeted gene knockout. Targeted knockout wheat with foreign DNA is generated in the T0 generation. Ultimately, the foreign DNA can be segregated by selfing and crossing. For example, researchers created a triple-knockout mutant of TaQsd1 via Agrobacterium delivered CRISPR/Cas9. The mutant was then crossed with wild-type wheat plants, producing transgene-free triple-recessive TaQsd1 mutants that exhibited longer seed dormancy (Abe et al., 2019). Similarly, a marker free wheat mutant was obtained among the offspring of T0 plants (Wang et al., 2017).
Sometimes, vectors are not integrated into the genome; instead, they may transiently express their encoded genome editing elements to knock out genes. A targeted gene-modified plantlet without foreign DNA is generated in the T0 generation. This approach has been reported in wheat for the first time. Researchers delivered vectors containing CRISPR/Cas9 elements into wheat callus through particle bombardment; the plantlet was subsequently regenerated without antibiotic selection. This transient expression-based CRISPR/Cas9 system produced transgene-free, homozygous mutants (Zhang et al., 2016). In addition, transgene-free wheat carrying nucleotide substitutions have been generated by transiently expressing CBEs or ABEs (Zong et al., 2017; Li et al., 2018).
DNA free genome-edited wheat plants have been generated. Though the editing efficiency was lower, the specificity was higher than with a vector-based system (Zhang et al., 2016). Moreover, nCas9-PBE mRNA and sgRNA were transcribed in vitro and delivered into immature
wheat embryos. DNA-free base editing at TaALS-P174 was obtained, endowing wheat with resistance to the herbicide nicosulfuron (Zhang et al., 2019). In addition, Cas9 can be expressed in vitro and assembled with the sgRNA into a Cas9/sgRNA ribonucleoprotein, which is delivered into immature wheat embryos by particle bombardment. The ribonucleoprotein cleaves the target site immediately and is quickly degraded, generating DNA-free edited wheat (Liang et al., 2017). The final CRISPR-edited DNA-free products are similar to natural and artificial mutants, which are not subject to GMO regulations. We believe that this is the direction of future breeding , and it will play a vital role in realizing sustainable agriculture in future.
Future Perspectives: The development of CRISPR/Cas9 technology has been extensively used in wheat genome editing. This technology permit multiplex genome engineering, which has enabled the production of loss-of function triple wheat mutants; thus, it is a powerful tool for introducing desired traits conferred by a loss-of function mutation into commercial cultivars through NHEJ. As additional genes required for genic male sterility are identified, CRISPR/Cas9- mediated disruption of these genes will enable the rapid production of male-sterile wheat. This represents a promising method for manipulating recessive sterility genes to capture heterosis in wheat.
CRISPR-mediated precise genome editing is a useful means to achieve these targeted substitutions and replacements by modifying endogenous genes without introducing linkage drag; it can also introduce new alleles (segregating as a single locus) into a predetermined genomic site. Thus, this approach could accelerate the breeding process. Nowadays, CRISPR-mediated precise genome editing is a useful means to achieve these targeted substitutions and replacements by modifying endogenous genes without introducing linkage drag; it can also introduce new alleles (segregating as a single locus) into a predetermined genomic site. Thus, this approach could accelerate the breeding process.
Trans-generational CRISPR/Cas9 activity has been used to modify multiple target sites in tomato and wheat (Rodriguez-Leal et al. 2017; Wang et al. 2018). This recommends that valuable, desired phenotypes in elite wheat germplasm, which are recalcitrant to transformation, could be induced by crossing with lines carrying CRISPR/Cas9 elements. In addition, wheat genes have been successfully edited through pollination using CRISPR/Cas9-transgenic maize as a haploid inducer (Budhagatapalli et al., 2020).
Such haploid induction-mediated genome editing would not only reduce the genotype dependence on site-specific mutagenesis in wheat, but also provide a path to produce transgene-free gene-edited inbred wheat lines. Collectively, these technologies will accelerate wheat breeding. Some studies have reported that although CRISPR/Cas9 can cleave a target site, sometimes it also cleaves sites with a few mismatches to the target site. This off target effect is a major concern in gene therapy, but this issue might not be a barrier in plant biotechnology. The putative off-target mutation could be eliminated through back-crossing or crossing with wild-type plants. Moreover, it is advisable to design target sites using web-based tools to reduce off-target mutations by leveraging computation.
Combined with other achievements, including the production of high quality genome sequences and improved transgenic methods, CRISPR and CRISPR-based genome editing will bring functional genomics and rational design-based molecular breeding of polyploid wheat to the forefront of wheat biology. We believe that transgene-free, gene edited wheat will play a critical role in addressing environmental issues while promoting sustainable agriculture. Significantly, it is not a replacement for traditional breeding; it is just one of the methods advancing wheat breeding programs and accelerating wheat biology.
REFERENCES
Abe F, Haque E, Hisano H, Tanaka T, Kamiya Y, Mikami M, Kawaura K, Endo M, Onishi K, Hayashi T et al (2019). Genome-edited triple-recessive mutation alters seed dormancy in wheat. Cell Rep. 28(5): 1362–1369.
Bibikova, M.; Golic, M.; Golic, K.G.; Carroll, D. (2002). Targeted chromosomal cleavage and mutagenesis in Drosophila using zinc-finger nucleases. Genetics. 161: 1169–1175.
Bortesi L, Zhu C, Zischewski J, Perez L, Bassie L, Nadi R, Forni G, Lade SB, Soto E, Jin X et al (2016). Patterns of CRISPR/Cas9 activity in plants, animals and microbes. Plant Biotechnol. J. 14: 2203–2216.
Budhagatapalli N, Halbach T, Hiekel S, Bu¨ chner H, Mu¨ uller A, Kumlehn J. (2020). Site-directed mutagenesis in bread and durum wheat via pollination by cas9/guide RNA-transgenic maize used as haploidy inducer. Plant Biotechnol. J. 18:2376–2378.
Christian, M.; Cermak, T.; Doyle, E.L.; Schmidt, C.; Zhang, F.; Hummel, A.; Bogdanove, A.J.; Voytas, D.F. (2010). Targeting DNA double-strand breaks with TAL effector nucleases. Genetics, 186: 757–761.
Cohen-Tannoudji, M.; Robine, S.; Choulika, A.; Pinto, D.; El Marjou, F.; Babinet, C.; Louvard, D.; Jaisser, F. (1998). I-SceI-induced gene replacement at a natural locus in embryonic stem cells. Mol. Cell. Biol. 18: 1444–1448.
Cong L, Ran FA, Cox D, Lin S, Barretto R, Habib N, Hsu PD, Wu X, Jiang W, Marraffini LA et al (2013) Multiplex genome engineering using CRISPR/Cas systems. Science 339: 819–823.
Cong, L.; Ran, F.A.; Cox, D.; Lin, S.; Barretto, R.; Habib, N.; Hsu, P.D.; Wu, X.; Jiang, W.; Marraffini, L.A. (2013). Multiplex genome engineering using CRISPR/Cas systems. Science. 339: 819–823.
Ding D, Chen K, Chen Y, Li H, Xie K (2018). Engineering introns to express RNA guides for Cas9- and Cpf1-mediated multiplex genome editing. Mol. Plant, 11, 542.
Endo M, Mikami M, Toki S (2015) Multigene knockout utilizing off target mutations of the CRISPR/Cas9 system in rice. Plant Cell Physiol. 56:41–47.
GAO C (2018). The future of CRISPR technologies in agriculture. Nat Rev Mol Cell Biol. 19:275.
Gibson DG, Young L, Chuang R-Y, Venter JC, Hutchison Iii CA, Smith HO (2009). Enzymatic assembly of DNA molecules up to several hundred kilobases. Nat Methods 6: 343.
He Y, Zhao Y (2020) Technological breakthroughs in generating transgene-free and genetically stable CRISPR-edited plants. aBIOTECH. 1: 88–96.
Hsu, P.D.; Lander, E.S.; Zhang, F. (2014). Development and applications of CRISPR-Cas9 for genome engineering. Cell. 157: 1262–1278.
Jinek, M.; Chylinski, K.; Fonfara, I.; Hauer, M.; Doudna, J.A.; Charpentier, E. A (2012). Programmable dual-RNA–guided DNA endonuclease in adaptive bacterial immunity. Science. 337: 816–821.
Jun Li, Yan Li, Ligeng Ma (2021). Recent advances in CRISPR/Cas9 and applications for wheat functional genomics and breeding. aBIOTECH. 2: 375–385
Karimi M, Depicker A, Hilson P (2007). Recombinational cloning with plant gateway vectors. Plant Physiol. 145: 1144–1154.
Kumar, A., Kapoor, P., Chunduri, V., Sharma, S., and Garg, M. (2019). Potential of aegilops sp. for improvement of grain processing and nutritional quality in wheat (Triticum aestivum L.). Front. Plant Sci. 10:308.
Li C, Zong Y, Wang Y, Jin S, Zhang D, Song Q, Zhang R, Gao C (2018). Expanded base editing in rice and wheat using a Cas9-adenosine deaminase fusion. Genome Biol. 19: 59.
Liang Z, Chen K, Li T, Zhang Y, Wang Y, Zhao Q, Liu J, Zhang H, Liu C, Ran Y. (2017). Efficient DNA-free genome editing of bread wheat using CRISPR/Cas9 ribonucleoprotein complexes. Nat Commun. 8: 14261.
Liu, M.; Rehman, S.; Tang, X.; Gu, K.; Fan, Q.; Chen, D.; Ma, W. (2019). Methodologies for improving HDR efficiency. Front. Genet. 9: 691.
Tofazzal Islam, Pankaj K. Bhowmik and Kutubuddin A. Molla (2020). CRISPR-Cas Methods, Springer Protocols Handbooks Springer Science+ Business Media, LLC, Part of Springer Nature.
Mali P, Yang L, Esvelt KM, Aach J, Guell M, DiCarlo JE, Norville JE, Church GM (2013). RNA-guided human genome engineering via Cas9. Science. 339: 823–826
Molla KA, Yang Y (2019). Predicting CRISPR/ Cas-induced mutations for precise genome editing. Trends Biotechnol. 38:136.
Nadolska-Orczyk A, Rajchel I, Orczyk W, Gasparis S (2017). Major genes determining yield-related traits in wheat and barley. Theor Appl Genet. 130: 1081–1098
Nalam VJ, Alam S, Keereetaweep J, Venables B, Burdan D, Lee H, Trick HN, Sarowar S, Makandar R, Shah J. (2015). Facilitation of Fusarium graminearum Infection by 9-Lipoxygenases in Arabidopsis and Wheat. Mol Plant Microbe Interact. 28: 1142-52.
Petersen G, Seberg O, Yde M, Berthelsen K (2006). Phylogenetic relationships of Triticum and Aegilops and evidence for the origin of the A, B, and D genomes of common wheat (Triticum aestivum). Mol Phylogenet Evol. 39(1): 70–82.
Rodriguez-Leal D, Lemmon ZH, Man J, Bartlett ME, Lippman ZB (2017). Engineering quantitative trait variation for crop improvement by genome editing. Cell 171(2): 470–480.
Shan Q, Wang Y, Li J, Zhang Y, Chen K, Liang Z, Zhang K, Liu J, Xi JJ, Qiu JL(2013). Targeted genome modification of crop plants using a CRISPR-Cas system. Nat. Biotechnol. 31: 686-8.
Simons KJ, Fellers JP, Trick HN, Zhang Z, Tai YS, Gill BS, Faris JD (2006). Molecular characterization of the major wheat domestication gene Q. Genetics. 172: 547-55.
Slavin, J.L.; Lloyd, B (2012). Health benefits of fruits and vegetables. Adv. Nutr. 3: 506–516.
Su Z, Hao C, Wang L, Dong Y, Zhang X (2011). Identification and development of a functional marker of TaGW2 associated with grain weight in bread wheat (Triticum aestivum L.). Theor Appl. Genet. 122: 211-23.
Sun, Y.; Zhang, X.;Wu, C.; He, Y.; Ma, Y.; Hou, H.; Guo, X.; Du, W.; Zhao, Y.; Xia, L. (2016). Engineering herbicide-resistant rice plants through CRISPR/Cas9-mediated homologous recombination of acetolactate synthase. Mol. Plant. 9: 628–631.
Uauy C, Wulff B, Dubcovsky J (2017). Combining traditional mutagenesis with new high-throughput sequencing and genome editing to reveal hidden variation in polyploid wheat. Annu Rev Genet. 51: 435–454.
Upadhyay SK, Kumar J, Alok A, Tuli R (2013). RNA-guided genome editing for target gene mutations in wheat. G3 (Bethesda). 3: 2233-8.
Veraverbeke, W. S., and Delcour, J. A. (2002). Wheat protein composition and properties of wheat glutenin in relation to breadmaking functionality. Crit. Rev. Food Sci. Nutr. 42, 179–208.
Wang K, Liu H, Du L, Ye X (2017a) Generation of marker-free transgenic hexaploid wheat via an Agrobacterium-mediated co-transformation strategy in commercial Chinese wheat varieties. Plant Biotechnol. J. 15(5): 614–623.
Wang W, Pan Q, He F, Akhunova A, Chao S, Trick H, Akhunov E (2018a). Transgenerational CRISPR-Cas9 activity facilitates multiplex gene editing in allopolyploid wheat. Crispr. J. 1(1): 65–74.
Wang Y, Cheng X, Shan Q, Zhang Y, Liu J, Gao C, Qiu JL.(2014). Simultaneous editing of three homoeoalleles in hexaploid bread wheat confers heritable resistance to powdery mildew. Nat. Biotechnol. 32: 947-51.
Wang, Y. Q., Li, M., Guan, Y. B., Li, L., Sun, F. S., Han, J. P., et al. (2019). Effects of additional cysteine residue of avenin-like b protein by site-directed mutagenesis on dough properties in wheat (Triticum aestivum L.). J. Agric. Food Chem. 67, 8559–8572.
Xing HL, Dong L, Wang ZP, Zhang HY, Han CY, Liu B, Wang XC, Chen QJ (2014). A CRISPR/Cas9 toolkit for multiplex genome editing in plants. BMC Plant Biol. 14: 327
Yang H, Wu JJ, Tang T, Liu KD, Dai C (2017). CRISPR/Cas9-mediated genome editing efficiently creates specific mutations at multiple loci using one sgRNA in Brassica napus. Sci Rep. 7: 7489.
Zhang R, Liu J, Chai Z, Chen S, Bai Y, Zong Y, Chen K, Li J, Jiang L, Gao C (2019). Generation of herbicide tolerance traits and a new selectable marker in wheat using base editing. Nat Plants. 5: 480–485.
Zhang Y, Liang Z, Zong Y, Wang Y, Liu J, Chen K, Qiu J, Gao C (2016). Efficient and transgene-free genome editing in wheat through transient expression of CRISPR/Cas9 DNA or RNA. Nat Commun. 7: 12617.
Zhang Y, Pribil M, Palmgren M, Gao C (2020). A CRISPR way for accelerating improvement of food crops. Nat food. 1: 200–205.
Zong Y, Wang Y, Li C, Zhang R, Chen K, Ran Y, Qiu J, Wang D, Gao C (2017). Precise base editing in rice, wheat and maize with a Cas9-cytidine deaminase fusion. Nat Biotechnol. 35(5): 438–440.


