Regenix Super Speciality Laboratories Pvt.Ltd. (Affliated to University of Madras).
Apollo Lab Services, Chennai India.
Article Publishing History
Received: 25/06/2023
Accepted After Revision: 10/09/2023
Cancer is caused by carcinogens. Among these causative agents viruses are the traceable source. Oncoviruses have both DNA and RNA as genetic material. The constant evolution of these viruses allows them to survive and thrive. HPV, HCV,HBV, HH8, EBV, HTLV, MCP are a few known carcinogenic viruses. The pathway of establishment and proliferation of the virus follows the order of initiation, promotion, and progression. Various viral proteins play a mechanistic role in the conversion of a normal cell into a neoplasm. This study is a small attempt to learn the evolutionary patterns of oncoviruses.
Two common pathways of cancer are the suppression of p53 and Rb genes. This study looks at the viral proteins that perform this activity and looks at the similarity through the evolution patterns of the viral proteins. The niche of this study lies in the identification of possible targets that can collectively prevent viral cancers. Also in multiplexing targets for diagnosis. However , a common target identification from a evolutionary point of view was not established. The study however establishes that among the evolved viruses onco- proteins have similarity i.e a common pathway of evolution.
Virus, Cancer, Mechanism, Protein.
Shyam P, Subramaniam S, Subramaniam S, Mythili U. Oncogenic Viruses: A Comparative Evolutionary Study. Biosc.Biotech.Res.Comm. 2023;16(3).
Shyam P, Subramaniam S, Subramaniam S, Mythili U. Oncogenic Viruses: A Comparative Evolutionary Study. Biosc.Biotech.Res.Comm. 2023;16(3). Available from: <a href=”http://surl.li/lqobn“>http://surl.li/lqobn</a>
INTRODUCTION
Cancer is the collective name for diseases caused by the uncontrolled division of abnormal cells which later proliferate and spread to other organs. The aggregation of such cells in one place leads to a ‘tumor, ‘(National Institutes of Health 2007, Anderson and Simon 2020). The process by which a cell converts from a normal to a malignant cell is called carcinogenesis. The factor that causes this mutation that triggers a cell to become malignant is called as carcinogens. Carcinogens are of 3 major types’ chemical carcinogens (including those from biological sources), physical carcinogens, and oncogenic (cancer-causing) viruses. Among these oncogenic carcinogens depend on persistence and take a longer time to cause malignancy, (Anderson and Simon, 2020).
This makes viral cancers the most detectable and preventable form of cancer. Viruses contain DNA or RNA as genetic material. Among these viruses with single-stranded DNA are known to evolve rapidly ( Zur, 1991). Evolution is the mechanism employed by an organism to overcome a hostile environment.The year 1964 saw the discovery of viruses also as a cause of cancer. Viruses cause cancer either directly or by causing inflammation. The first of the oncogenic viruses to be discovered was Epstein-Barr Virus (EBV). The expression of proteins is a key step in the infection process and hence making immunosuppressed patients more suitable for viral cancers, (Tonseilo et al 2018 Tempera & Lieberman, (2021).
The key 7 types of viruses are proven to be oncogenic in nature namely. Viruses associated with causing cancer are EBV (Epstein–Barr virus), HPV (human papillomavirus), HTLV-1 (human T-lymphotropic virus 1), KHSV (Kaposi sarcoma-associated herpesvirus)/ HHV-8 (human herpesvirus 8), HCV (hepatitis C virus), HBV (hepatitis B virus) and MCPyV (Merkel cell polyomavirus). Among the above-mentioned HTLV-1, HCV and HBV are retroviruses. Constant assault leads to malignant transformation.2.2 million infections were attributed to cancer cases in 2018 worldwide which amounted to 13 % of the total cancers. 90% of the cases are caused by HPV, HBV, and HCV (Liao.,2006). Among the various diseases cancer is the highest death causing disease, accounting for almost 10 million deaths in 2020, (WHO 2022).
Among the above-mentioned viruses, most of them are DNA viruses and two of them are RNA viruses. Most RNA viruses are retroviruses that replicate via reverse transcriptase. EBV and KSHV are large DNA viruses that often cause solid tumors and lymphoid malignancies, (Charostad et al., 2020).
HPV and MCPyV are smaller DNA genomes in size and cause warts. Where oncogenic HPVs establish persistent infections in mucosal epithelia whereas MCPyV infects and likely persists latently in dermal fibroblasts. These small DNA oncogenic viruses promote tumorigenesis using relatively few multifunctional oncoproteins. HCV a retroviral virus has genetic material in a positive sense, HBV, on the other hand, is single a stranded RNA virus, that infects hepatocytes and causes chronic liver inflammation, liver cirrhosis, and hepatocellular carcinoma (HCC). HTLV-1 has RNA as genetic material and causes T-Cell lymphoma.
Infection of the above viruses is common but with viral cancers, there is no thumb rule that cancer progression needs the presence of virus. The general process of oncogenic viruses to cause cancer is chronic inflammation, immunosuppression, or environmental mutagens individually or in combination cause assault and helps with the progression to cancer. Several studies have been undertaken to prove that viruses are the root cause of cancer. Studies on both human and animal models have ascertained the same.
Viral mechanisms of inducing infection are direct and indirect mechanisms. Direct mechanisms involve. Insertion of viral oncogenes into host cells followed by enhancement of the proto-oncogenes in the host genes (Coffin et al., 1997) Indirect viral oncogenes activation occurs to triggering non–specific inflammation. It happens due to prolonged periods of inflammation. The direct method of infection mandates the presence of at least one virus in every tumor cell, expressing a minimum of one protein or RNA to help the cell become cancerous. Indirect tumor viruses are hypothesized to be lost from the tumor after the tumor has the sufficient mutation and attains hyperplasia (Morales-Sánchez & Fuentes-Pananá 2014 and Gaglia & Munger 2018).
Viral cancers are characterized by long latent periods of incubation. Cancers induced by viruses are part of the natural viral life cycle. RNA viruses have 3 distinct methods of transformation i.e., acute transformation, v-oncogene replication, and non-transforming replication. Oncoviruses often use pathways common pathways to induce tumorigenesis, some common pathways used by viruses to induce cancer are tumor suppressor pathways such as p53 and tumor necrosis–associated pathways (TRAFs), telomerase reverse transcriptase (TERT) and other pathways used are P13K-AKT-mTOR, JAK /STAT, NF-κB, β-catenin, MHC-1.The host DNA damage response is catalyzed and influenced by the viral proteins. In the study of the Kegg pathway, it was seen that the most common pathways of the above-mentioned p53 suppression, P13K-AKT-mTOR, and JAK /STAT pathways inhibit apoptosis ( Ozaki & Nakagawara, 2011), Brooks & Putoczki, 2022).
In this study, the general evolution of the oncogenic viruses is studied with the proteins involved in viral oncogenesis. Further, the evolution of the DNA viruses was explored, and finally the evolution of the proteins involved in the suppression of apoptosis through p53, P13k-AKT –mTOR, and JAK /STAT pathways. The study of these mechanisms shows a common route of infection by the oncoviruses. This study aims at understanding the common mechanisms of neoplasm induction by viruses and the evolutionary patterns of viral oncogenes involved in common mechanisms causing cancer.
MATERIAL AND METHODS
Retrival of whole genome structures from NCBI was carried out for – Epstein–Barr virus, Human T-lymphotrophic virus -1, Kaposi sarcoma-associated herpesvirus/ human herpesvirus8 , Human papillomaviruses, Merkel cell polyomavirus, Hepatitis B virus and Hepatitis C virus.Retrival of protein and gene sequences of the main viral onco-proteins from NCBI. Kegg pathway analysis:
and Pathways in cancer – Homo sapiens (human)- pathway number hsa 05200 was also studied.
Viral carcinogenesis- pathway number map05203 was analyzed to identify and select common pathways in cancer caused by Viruses and to identify the viral genes involved in the mechanism of carcinogenesis. Similarly, multiple sequence alignment of the chosen virus and viral proteins were aligned to see similarity using Clustal omega, Clustal W and Mega X. Construction of evolutionary tree was done by using Clustal omega, Clustal W and Mega X based on neighbor joining method.
RESULTS AND DISCUSSION
The phylogenetic analysis of the proteins that induce oncogenesis is depicted in the below phylogenetic tree. The analysis results from Clustal omega and Clustal W are given below. Cancer causing viruses have both RNA and DNA as genetic material. Hence to analyze a common pattern of infection and carcinogenesis using keg analysis the main viral proteins were identified and taken for study. The sequences taken for study are listed in the table below.
Table .1 List of Viral proteins taken for the study.
| ENTRY | ENTRY NAME | PROTEIN
NAME |
GENE
NAME |
ORGANISM | LENGTH |
| P03211 | EBNA_EBVB9 | EPSTEIN- BARR NUCLEAR
ANTIGEN 1 |
EBNA, BKRF1 | EPSTEIN-BARR VIRUS (STRAIN B95-8) | 641AA |
| P12977 | EBNA3_EBVB9 | EPSTEIN- BARR
NUCLEAR ANTIGEN 1 |
EBNA3 | EPSTEIN-BARR VIRUS (STRAIN B95-8) | 944AA |
| A0A3R5XA97 | A0A3R5XA97_EBVG | EBNA-3A | EBNA- 3A | EPSTEIN BARR
VIRUS (STRAIN GD1) (HHV-4) |
925 AA |
| A0A075FDV6 | A0A075FDV6_EBVG | EBNA-3B | EBNA- 3B | EPSTEIN BARR VIRUS (STRAIN
GD1) (HHV-4) |
892AA |
| A0A0C7P4C5 | A0A0C7P4C5_EBVG | EBNA-3C | 3BNA-
3C |
EPSTEIN BARR
VIRUS (STRAIN GD1) (HHV-4) |
992AA |
| K9USE8 | K9USE8_EBVG | EBNA-LP | EBNA- LP | EPSTEIN BARR VIRUS (STRAIN
GD1) (HHV-4) |
506AA |
| Q67978 | Q67978_HBV | CAPSID
PROTEIN |
CORE | HEPATITIS B
VIRUS (HBV) |
212AA |
| Q89656 | Q89656_HBV | CAPSID
PROTEIN |
PRE-
C/CORE |
HEPATITIS B
VIRUS (HBV) |
212AA |
| P03141 | HBSAG_HBVA3 | LARGE ENVELOPE PROTEIN | S | HEPATITIS B VIRUS GENOTYPE A2SUBUNIT ADW2(STRAIN RULTER 1979)
(HBV-A) |
400AA |
| Q9QMN6 | Q9QMN6_HBV | LARGE ENVELOPE
PROTEIN |
S | HEPATITIS B VIRUS (HBV) | 400AA |
| Q9QMN7 | Q9QMN7_HBV | PROTEIN P | P | HEPATITIS B
VIRUS (HBV) |
843AA |
| P06463 | VE6_HPV18 | PROTEIN_E6 | E6 | HPV TYPE 18 | 158AA |
| P03126 | VE6_HPV16 | PROTEIN_E6 | E6 | HPV TYPE 16 | 158AA |
| P04014 | VE1_HPV11 | REPLICATION
PROTEIN E1 |
E1 | HPV TYPE 11 | 649AA |
| P27223 | VE2_HPV42 | REGULATORY
PROTEIN E2 |
E2 | HPV TYPE 31 | 398AA |
| Q705H5 | VL2_HPV43 | MICROCASPID PROTEIN L2 | L2 | HPV TYPE 45 | 463AA |
| Q80915 | VE1_HPV44 | REPLICATION
PROTEIN E1 |
E1 | HPV TYPE 45 | 643AA |
| P17387 | VE7_HPV31 | PROTEIN E7 | E7 | HPV TYPE 31 | 98AA |
| P06427 | VE6_HPV33 | PROTEIN E6 | E6 | HPV TYPE 18 | 149AA |
| A0A0P0ERM8 | A0A0P0ERM8_HPV34 | REGULATORY
PROTEIN E2 |
E2 | HPV TYPE 31 | 345AA |
| A0A6M3S242 | A0A6M3S242_HPV35 | REPLICATION
PROTEIN E1 |
E1 | HPV TYPE 16 | 637AA |
| T2A6T1 | T2A6T1_HPV39 | PROTEIN_E6 | E6 | HPV TYPE 16 | 158AA |
| W5RS47 | W5RS47_HPV45 | PROTEIN E6 | E6 | HPV45 | 158AA |
| C9EF17 | C9EF17_HPV51 | MINOR CASPID
PROTEIN L2 |
L2 | HPV 51 | 469AA |
Figure: 1 Clustal omega based tree for viral evolution
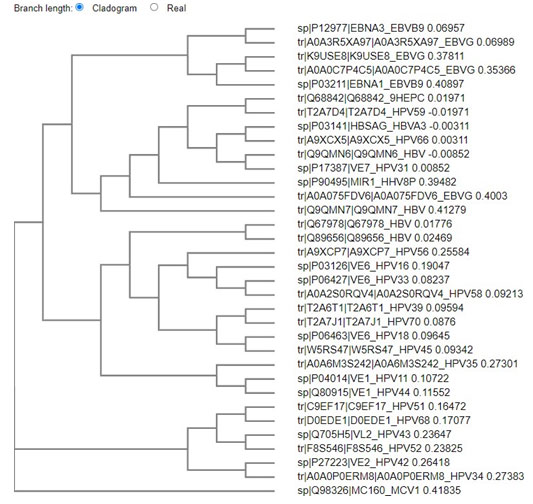
Figure: 2 Clustal W based phylogenetic tree
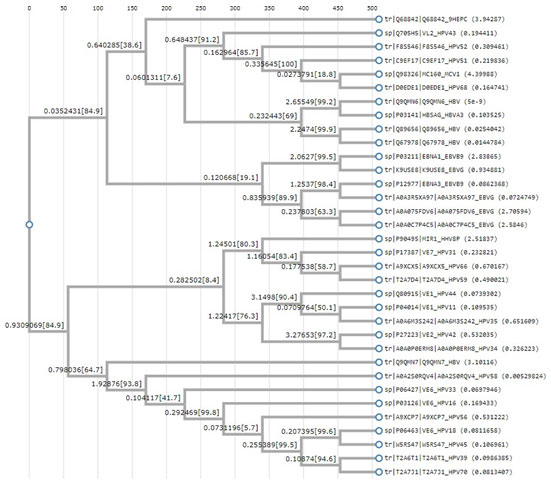
The above tree shows vast evolution of the viral proteins. HTLV is the earliest known of the carcinogenic viruses. The evolution of the viruses based on the viral proteins takes place as two branches. The evolution takes place in clusters with the proteins of one virus most closely related to the other onco- proteins of the same virus. Although it is seen the EBV and HPV have common points of evolution. HBV and HPV viral protiens also are seen to have common point of origin. One marked point of similarity is between the protein of HHV 8 and HPV 31, both owing to be cancer causing viruses of the epithelium. Hence from this analysis it can be concluded that no common primers or multiplexing technique can be employed to identify the cancer causing viruses. Since viruses are both of DNA and RNA in nature , the next phase of screening was based on the DNA viruses.
Phylogenetic analysis of the DNA viruses : The results from the phylogenetic analysis are depicted from Mega X.
Figure: 3 Phylogeny of DNA viruses using Mega X
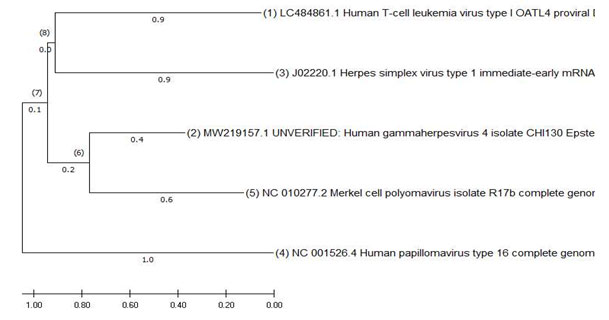
The interpretation of this tree was done by neighbor joining method.Evolutionary analyses were conducted in MEGA X .The Maximum likelihood method infers the topology of the tree. HPV is the most diverged of the viruses in studied group.1,2,3 denotes the divergence patterns of the taxa .Order of evolution is from the tree is HTLV-1 , the next divergence in the codons evolution is EBV. The next evolution is KSHV and the arm length . The most divergent is HPV which comes next in the series . The last of the evolution is MCV.
The distances show evolution and variations in the codons. The HTLV-1 and EBV deal with lymphomas. The HH8 and MCV deal with skin cancers. Whereas, HPV deals with a group of specialized cells of the cervix, anus, penile etc., forming carcinomas in the epithelium and metastases into the squamous cells. The evolution and divergence shows the increased ability of the cancers to establish in the specialized regions.
Phylogenetic analysis of proteins that trigger the MAP-Kinase and JAK –STAT pathway:MAP Kinase pathway –Mitogen activated protein kinase pathway is involved in several organized and linked signaling cascades which often involve oncogenesis , tumor progression and drug resistance. MAPK are a group of kinases whose activities are altered for cancer development. This makes the MAP-kinase pathway a reliable target for cancer therapy.
JAK –STAT pathway – plays a catalytic role in tumor progression .The pathway triggers tumor growth /metastasis or as an immune system modulator. The over stimulation of JAK/STAT pathway proteins leads to proliferation, progression, survival and metastasis of cancer cells.
Figure: 4 Phylogeny of MAP-Kinase and JAK –STAT pathway using Mega X

The evolutionary history was inferred by using the Maximum Likelihood method .This analysis involved 4 amino acid sequences. There were a total of 1133 positions in the final dataset. Evolutionary analyses were conducted in MEGA X (Kumar., et al .2018).The MAP-kinase and JAK –Stat pathway promotes immortalization through a series of pathways. The proteins in the viruses coding for the initiation of these pathways were compared to see the evolutionary pathway. Based on the distances the HPV E7 and TAX of HTLV1 are the most divergent. The LMP1 of the EBV and GPCR of KSHV are the intermediates in this evolutionary process.
Phylogenetic analysis of proteins that block the p53 activity: P53 is attributed to promotion of apoptosis. p53 responds to factors such as DNA damage and promotes a cell cycle arrest by accumulation in the nucleus. If the DNA damage is irreparable then p53 promotes cell death.
Figure: 5 Phylogeny of proteins that block p53

Neighbor joining method was used for this phylogenetic analysis.Evolutionary analyses were conducted in MEGA X (Kumar., et al .,2018).
The inactivation of p53 is one of the major steps in the establishment of carcinogenisis. The constant presence of the virus and multiplication helps in the establishment of carcinogenesis. This tree indicates EBNA-1 and E6 of HPV are most distantly related. KSHV , LANA and TAX1 of HTLV1.
The evolution and mutation the species undergo is for the adaptation purpose. HPV is the most evolved of the studied viruses and this evolution has led to the many subtypes. Viral cancers take a long time to establish and convert to neoplasm and carcinogenesis. This is the key factor to curing the viral cancers. Viral cancer often manipulates pathways that coincide with the hall marks of cancer. The proteins that trigger and establish carcinogenesis have undergone evolution indicating them as the sources for detection / manipulation for treatment. Viral infections often provide the entropy that defines the genetic and phenotypic variation, hence manifesting itself as the driving force of cancer evolution (Tempera & Lieberman 2021).
Among the viruses EBV and KSHV are large DNA viruses who often cause solid tumours and lymphoid malignancies. HPV and MCPyV are smaller DNA genomes in size and cause wrats. Where oncogenic HPVs establish persistent infections in mucosal epithelia whereas MCPyV infects and likely persists latently in dermal fibroblasts. These small DNA oncogenic viruses promote tumorigenesis using relatively few multifunctional oncoproteins. HCV a reteroviral virus has genetic material as a positive-sense, HBV on the other hand are single stranded RNA virus,both infect hepatocytes and cause chronic liver inflammation, liver cirrhosis and hepatocellular carcinoma (HCC) ( Zuckerman, et al., 1997).
HTLV-1 is a human oncogenic retrovirus that affects T cells and can initiates adult T cell lymphoma Deregulation of p53 and RB pathways is common mechanisms of most oncogenic viruses. The oncoproteins suppress p53 and RB by methods of degradation, inactivation, disassociation or repression (Tornesello, et al.,2018). The viruses have use different mechanisms for suppression, the proteins taken for the study are the main proteins that initiate the pathway. However the RB sequence is not part of all the viruses under study and mainly is only a part of the cancers involving epithelium.
CONCLUSION
The evolutionary study gives an insight to the mechanisms of evasion followed by the viruses and variations it undergoes over the years to thrive and hence providing an insight to treatment / prevention of the cancers. Form this study it can be concluded that a common target for viral cancers cannot be chosen. However, cancer causing viruses of similar organs ( eg. Epithelium) seem to have a common point of origin. Further studies with the mechanisms of these proteins may give us an insight into preventable targets for viral cancers.
REFERENCES
Anderson NM and M C Simon The tumor microenvironment Curr Biol 2020 Aug 17;30(16):R921-R925.
Brooks, A. J., and Putoczki, T. (2022). JAK-STAT Signalling Pathway in Cancer. Cancers, 12(7), 1971
Charostad, J., Nakhaie, M., Dehghani, A., & Faghihloo, E. (2020). The interplay between EBV and KSHV viral products and NF-κB pathway in oncogenesis. Infectious agents and cancer, 15, 62. https://doi.org/10.1186/s13027-020-00317-4
Coffin JM, Hughes SH, Varmus HE, editors. Retroviruses. Cold Spring Harbor (NY): Cold Spring Harbor Laboratory Press; 1997. Oncogenesis.
Gaglia, M. M., & Munger, K. (2018). More than just oncogenes: mechanisms of tumorigenesis by human viruses. Current opinion in virology, 32, 48–59. https://doi.org/10.1016/j.coviro.2018.09.003.
Kumar S., Stecher G., Li M., Knyaz C., and Tamura K. (2018). MEGA X: Molecular Evolutionary Genetics Analysis across computing platforms. Molecular Biology and Evolution 35:1547-1549.
Liao J. B. (2006). Viruses and human cancer. The Yale journal of biology and medicine, 79(3-4), 115–122.
Morales-Sánchez, A., & Fuentes-Pananá, E. M. (2014). Human viruses and cancer. Viruses, 6(10), 4047-4079.
National Institutes of Health (US); Biological Sciences Curriculum Study. NIH Curriculum Supplement Series [Internet]. Bethesda (MD): National Institutes of Health (US); 2007. Understanding Cancer. Available from: https://www.ncbi.nlm.nih.gov/books/NBK20362/
Ozaki, T., & Nakagawara, A. (2011). Role of p53 in Cell Death and Human Cancers. Cancers, 3(1), 994–1013.
Tempera, I., & Lieberman, P. M. (2021). Oncogenic viruses as entropic drivers of cancer evolution. Frontiers in virology, 1, 753366.
Tornesello, M. L., Annunziata, C., Tornesello, A. L., Buonaguro, L., & Buonaguro, F. M. (2018). Human Oncoviruses and p53 Tumor Suppressor Pathway Deregulation at the Origin of Human Cancers. Cancers, 10(7), 213. https://doi.org/10.3390/cancers10070213.
World Health Organization (2022) https://www.who.int/news-room/fact-sheets/detail/cancer
Zuckerman AJ. Hepatitis Viruses. In: Baron S, editor. Medical Microbiology. 4th edition. Galveston (TX): University of Texas Medical Branch at Galveston; 1997. Chapter 70. Available from: https://www.ncbi.nlm.nih.gov/books/NBK7864/
Zur Hausen H. (1991). Viruses in human cancers. Science (New York, N.Y.), 254(5035), 1167–1173.


