Department of Biotechnology, Jamia Millia Islamia, New Delhi, India.
Corresponding author email: kdev@jmi.ac.in
Article Publishing History
Received: 15/10/2021
Accepted After Revision: 05/02/2022
Type 2 diabetes mellitus (T2DM) is a chronic disorder characterized by pancreatic beta-cell dysfunction and insulin resistance. The present study was designed to understand the association of genetic variations in the JAZF1 gene with T2DM in the Indian population. The polymorphic study was conducted by PCR-RFLP methods. Further, the biochemical parameters were collected for statistical analysis on the semi-structured questionnaire, and correlation with the polymorphism was done by using SPSS software.
The significant differences were observed between T2DM cases and controls in triglycerides, HbA1c, T-cholesterol, LDL-C, BMI, systolic and diastolic BP, PPG, FPG, while no significant differences were observed in HDL-C, WHR. Our results suggested that the JAZF1 rs864745 variant is significantly associated with T2D among the Indian population. The present study concludes that the association of genetic variations and biochemical factors play a vital role in T2DM risk and its prevalence.
Hyperglycaemia, Indian Population, JAZF1, Type 2 Diabetes Mellitus.
Goyal Y, Verma A. K, Dev K. On the Efficacy of the Gene, Juxtaposed with Another Zinc Finger Protein 1 (JAZF1) in the Development of Type 2 Diabetes Mellitus among Indians. Biosc.Biotech.Res.Comm. 2022;15(1).
Goyal Y, Verma A.K, Dev K. On the Efficacy of the Gene, Juxtaposed with Another Zinc Finger Protein 1 (JAZF1) in the Development of Type 2 Diabetes Mellitus among Indians. Biosc.Biotech.Res.Comm. 2022;15(1). Available from: <a href=”https://bit.ly/2U8EBeg”>https://bit.ly/2U8EBeg</a>
Copyright © This is an Open Access Article distributed under the Terms of the Creative Commons Attribution License (CC-BY). https://creativecommons.org/licenses/by/4.0/, which permits unrestricted use distribution and reproduction in any medium, provided the original author and sources are credited.
INTRODUCTION
During the last couple of decades, the prevalence of diabetes has increased drastically all over the world and now diabetes disease has become a worldwide public health problem. According to the International Diabetic Federation (IDF-2017), a total of 8.8% of the World’s population was suffering from diabetes and this population of 425 million is estimated to further increase to 628.6 million people by (2045). Diabetes has established itself as one the fastest growing disease in humans and has become an epidemic with a 48% increase in the last 30 years. Its prevalence has continuously increased in the adults 20-79 years’ age group from 151 million in (2000), to 285 million in (2009) to 382 million in 2013 and 424.9 million in (2017) (Zimmet 2017; Brussels and Belgium 2019).
In India, 72.9 million people are suffering from diabetes and by (2045) this patients count is expected to be 134.3 million. Diabetes accounts for high morbidity and mortality due to complications like renal failure, amputations, cardiovascular disease, and cerebrovascular events (Schulze et al. 2004; Park et al. 2020). Nearly half of those affected are undiagnosed. Furthermore, among all major ethnic groups, Asian Indians have one of the highest incidences of pre-diabetes and diabetes, and the transition from pre-diabetes to diabetes occurs more quickly in this community (Anjana et al. 2011). Long-term diabetes has major problems, some of which are fatal (Alam et al. 2021).
The successful discovery of common (SNPs) contributing to diabetes susceptibility has been made possible by technological advances in molecular biology. Genome-wide techniques, such as (GWAS), have found statistically significant links between certain genetic regions and T2DM risk (Basile et al. 2014). In humans, several JAZF1 single nucleotide polymorphisms (SNPs) have been linked to T2DM and IR-related disorders. The replacement of Asparagine for Alanine is caused by a well-known missense mutation of rs1635852 (C to T substitution).
The rs1635852-T risk allele in JAZF1 was linked to T2DM in meta-analysis research involving approximately 1 million participants (Fogarty et al. 2013; Mahajan et al. 2014). Patients with the T risk allele had lower JAZF1 mRNA expression and higher protein complex binding (Fogarty et al. 2013). It’s worth noting that T risk allele carriers had lower insulin secretion due to transcriptional suppression of PDX1, a key transcription regulator for beta-cell formation and regeneration (Zhu et al. 2017).
Another explanation is that rs1635852 mutations cause insulin exocytosis to be reduced by binding to miR-96 (Li et al. 2016). However, not only is there a scarcity of information about the rs864745 variant of JAZF1, but its relationship to T2DM in the Indian population is also unknown. The goal of this case-control study was to see if the JAZF1 variation polymorphism (rs864745) was associated with an increased risk of T2DM. Thus, the present study analyses the significance of the association of genetic of JAZF1 gene polymorphism in the risk of development T2DM as well as the correlation with the clinical biochemical parameters in the Indian population.
MATERIAL AND METHODS
The present study includes a total of 300 persons and was conducted at Medical Biotechnology Laboratory, Dept. of Biotechnology, Jamia Millia Islamia (JMI) (A Central University), New Delhi, amongst these 300 individuals, 200 individuals were newly diagnosed T2DM cases the and remaining 100 healthy controls. Patients with T2DM and healthy controls were chosen on basis of the inclusion and exclusion criteria. This study was performed only after the due approval of the institutional ethical committee, JMI, New Delhi.
Patients included in the present study were examined and collection of samples was done only after informed consent from all study participants. Information of patients was taken in standardized questionnaires. Isolated DNA was amplified to determine JAZF1 rs864745 (A/G) genotype by a particular set of primers; forward primer: 5’- GAGCCATATAAGTGATGCTCAAA-3’ and reverse primer: 5’- GGTTGTCAGGCTTTCCATGT-3’ using thermal cycler.
The amplified DNA product of 378 bp was viewed with an ultraviolet (UV) transilluminator. JAZF1 rs864745 (A/G) polymorphism was identified by the SSPI restriction enzyme identifying the sequence of DNA. DNA band showed 378 bp A allele- uncut, G allele- 338 + 40 bp. Frequencies of genotypes between patients (cases) and healthy individuals were assessed by Chi-square test and those values which were <0.05, were evaluated by Fisher-exact test (Gong et al. 2021).
The link between JAZF1 rs864745 (A/G) and T2DM risk was projected by calculating the odds ratios (OR) value with 95% confidence intervals (CI). P-value <0.05 considered significant (Morris 2018). The study has been conducted only after the due clearance and approval from the Ethics committee of Jamia Millia Islamia (vide Proposal No. 26/11/273/JMI/IEC/2019). As part of the mandatory standardized ethical norm, written informed consent was taken from the person before inclusion in the research work.
RESULTS AND DISCUSSION
Genotype and allele frequencies of JAZF1 gene polymorphism in patients and controls: Table 3.1 illustrates genotypes and alleles frequencies, odds ratios, 95% confidence intervals, and P values for the three ‘JAZF1gene polymorphism among T2D patients and controls. JAZF1 (rs864745) showed a high percentage of Homozygous mutant GG 55 (27.5%) in patients as compared to controls GG 10 (10%A). A 2% difference was observed in the case of heterozygous AG in patients (28%) as compared to control (26%). A low percentage of homozygous AA (44.5) was observed in patients as compared to controls (64%). The odds ratio of JAZF1 genotype AG (heterozygous) and GG (Mutant Homozygous) with AA (wild homozygous), 1.54 (0.88-2.72), and 3.95 (1.87-6.70) were observed, respectively. We observed a significant difference in the frequency of risk allele ‘G’ among patients and controls (p<0.0001).
Table 1. Genotypic and allelic frequencies of JAZF1 gene polymorphism
among T2D cases and controls.
| Gene/SNP ID | Genotype/Allele | Cases
(n=200) |
Control (n=100) | Odd Ratio
(95% CI) |
P-value |
|
JAZF1 (rs864745) |
AA | 89 (44.5%) | 64 (64%) | Ref | Ref |
| AG | 56 (28%) | 26 (26%) | 1.54 (0.88 – 2.72) | 0.12 | |
| GG | 55 (27.5%) | 10 (10%) | 3.95 (1.87 – 6.70) | <0.001* | |
| P-value < 0.001* | |||||
| A (Normal allele) | 234 (58.5%) | 154 (77%) | 2.37 (1.61 – 3.48) | <0.0001* | |
| G (Risk allele) | 166 (41.5%) | 46 (23%) | |||
Note: * = P-value < 0.05 considered significant.
The frequencies, odds ratios, and P-values of the JAZF1 (rs864745) genotypes among T2D patients and control subjects under dominant and recessive models
Table 3.2 shows the frequencies, OR and, p-values of dominant and recessive models of JAZF1 (rs864745) among T2D patients and controls. There is a significant difference was observed between the two groups under the dominant and recessive models (p-value < 0.001).
Table 2. Frequencies, OR, and p-values of dominant and recessive models
of JAZF1 (rs864745) among T2D patients and controls
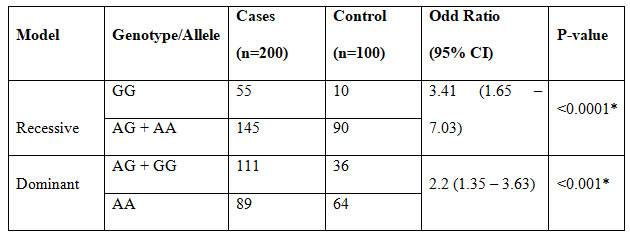
Note: * = P-value < 0.05 considered significant.
Comparative analysis of the Biochemical parameters in T2D cases and controls
Diabetes is a multifactorial disorder and along with genotype, different factors come into play to develop this condition. Tables 3 illustrates comparative analysis of biochemical factors of T2D patients and controls in Delhi population. A significant association was observed in all levels among different JAZF1 genotype except HDL-C and WHR.
Table 3. Comparative analysis of clinicopathological parameters among T2D patients and controls.
| Factors | T2DM patients | Controls | P-values |
| Number | 200 | 100 | — |
| Age (in years) | 42.4 ± 9.3 | 40.7 ± 8.2 | |
| Triglycerides (mg/dl) | 347.7 ± 98.5 | 139.7 ± 6.1 | <0.001* |
| HbA1c | 7.9 ± 0.9 | 5.5 ± 0.7 | <0.001* |
| T-Cholesterol (mg/dl) | 242.6 ± 14.8 | 151.3 ± 19.1 | <0.001* |
| LDL-C (mg/dl) | 189.4 ± 28.7 | 104.3 ± 20.1 | <0.001* |
| HDL-C (mg/dl) | 47.1 ± 10.9 | 45.9 ± 9.1 | 0.180 |
| BMI (kg/m2) | 29.9 ± 4.9 | 25.1 ± 1.9 | <0.001* |
| Systolic BP (mmHg) | 144.9 ± 16.8 | 105.7 ± 9.9 | <0.001* |
| Diastolic BP (mmHg) | 101.9 ± 16.1 | 76.1 ± 11.2 | <0.001* |
| PPG | 229.8 ± 36.2 | 135.8 ± 4.9 | <0.001* |
| FPG (mg/dl) | 160.4 ± 25.7 | 89.8 ± 6.9 | <0.001* |
| WHR | 1.0 ± 0.2 | 1.0 ± 0.1 | 1.0 |
Note- Data presented as Mean ± SD; P-value (*) <0.05 considered significant.
HbA1c – haemoglobin A1c test, LDL – Low density lipoprotein, HDL – High-density lipoprotein, BMI – Body mass index, BP – Blood pressure, PPG – Postprandial plasma glucose, FPG – Fasting plasma glucose, WHR – Waist to Hip Ratio.
GWAS has been identified in more than 150 different loci associated with type 2 diabetes (Suzuki et al. 2019). Environmental factors are associated with T2DM onset which includes inactive/sedentary lifestyle, obesity, and stress (Adeghate et al. 2006). We analyzed various demographic and clinical parameters and significant differences were observed between T2DM patients and healthy controls in triglycerides, HbA1c, T-cholesterol, LDL-C, BMI, systolic and diastolic BP, PPG, FPG, while no significant differences were observed in parameters such as HDL-C, WHR, among T2DM patients and controls (Alam et al. 2021).
According to the CARRS (Centre for Cardio-metabolic Risk Reduction in South Asia) Study, the total prevalence of diabetes in three major cities in South Asia was 22.8 percent (21.5-24.1 percent), 25.2 percent (23.6-26.8 percent), and 16.3 percent (15.2-17.3 percent) (Deepa et al. 2015). Diabetes prevalence varies significantly depending on where you live (less in rural areas) and your socioeconomic status (less in low socio-economic stratum). Diabetes prevalence ranged from 3% in rural Jharkhand, east India, to 13.7 percent in metropolitan Tamil Nadu, south India, according to the ICMR-INDIAB research (Anjana et al. 2011).
Men (3.33 per 1000 per year) have been reported to have a faster rate of increase in diabetes prevalence than women (0.88 per 1000 per year) (Mishra and Khurana, 2011). The human JAZF1 gene has five exons and is found on chromosome 7p15.2. JAZF1 is a 243-amino-acid protein with a predicted molecular mass of 27 kDa. JAZF1 is made up of three zinc-finger domains with a repeating Cx(4)C2H and a ligand-binding domain (residues 341-583) (Koontz et al. 2001; Nakajima et al. 2004). Endometrial stromal tumors are linked to chromosomal abnormalities involving this gene (Koontz et al. 2001). Different protein isoforms are encoded by alternatively spliced variants, which have been described. Not all varieties, however, have been adequately characterized (Alam et al. 2021).
The human JAZF1 protein shares 90% homology with that of chimps, monkeys, mice, and pigs, implying that JAZF1 may have a comparable biological function in diverse species (Yang et al. 2015). In the present study, we examined the association of gene polymorphism in the JAZF1 gene to the risk of T2DM in the Indian population. JAZF1 rs864745 (A/G) variant association with T2DM has been reported by various studies among numerous populations (Koontz et al. 2001; Nakajima et al. 2004). Besides lifestyle and environmental risk factors, type 2 diabetes mellitus also has an established genetic predisposition (Zhang et al. 2019; Alam et al. 2021).
Our results suggested that the JAZF1 rs864745 variant is significantly associated with T2D among the Indian population. It was observed that GG genotype frequency was significantly higher in T2DM cases as compared to healthy controls. Allelic frequency of G allele was higher in T2DM cases in comparison to healthy controls. We found significant relations of JAZF1 polymorphism and T2DM risk under dominant and recessive models. Similar results were observed in studies conducted among Chinese and Iranian populations (Han et al. 2010; Soltanian et al. 2020; Alam et al. 2021).
The link between the rs864745 variation and T2DM, as well as the mechanism behind it, has been explored. The probability of developing T2DM was 2.32 times higher among Uyghurs with the rs864745-C risk allele (Song et al. 2015). Subjects with the rs864745-T risk allele, on the other hand, were found to have a lower risk of GDM (Stuebe et al. 2014). Based on mechanism, rs864745-T polymorphisms in the JAZF1 gene are linked to lower JAZF1 mRNA expression and insulin secretion (Grarup et al. 2008; Zano et al. 2020).
Increased fasting plasma insulin concentration is connected to rs864745-T polymorphisms, according to an autosomal genomic scan (Grarup et al. 2008). Several published research, on the other hand, have discovered a link between the rs864745 gene variant and T2DM-related illnesses. To begin, the rs864745-T variant of the JAZF1 gene is substantially linked to arteriolosclerosis in neuropathologic studies (Chou et al. 2013). The Saudi population with the JAZF1 rs864745-G risk allele had lower BMI and waist circumference (Alharbi et al. 2015).
The G-risk allele has also been linked to T2DM and lowered eGFR, which is consistent with lower JAZF1 gene expression in the peripheral blood of DN patients (Chen et al. 2013; Peng et al. 2017). Several studies have already been published that show a link between the JAZF1 (rs864745) mutation and T2DM, particularly in industrialized nations, but additional research is needed to use this gene as a biomarker (Zano et al. 2020). Environmental, metabolic, and genetic factors all play a role in the development of T2DM, according to previous research (Geng and Huang 2020).
CONCLUSION
The findings of the present study concluded that JAZF1 gene polymorphism was found to be associated with the risk of T2DM in the Indian population. Our study concludes association of genetic and biochemical factors plays a significant role in potential risk associated with the prevalence of T2DM and the JAZF1 gene may increase the severity of T2DM specifically in the Indian Population.
Conflict of Interests: Authors declare no conflicts of interests to disclose.
Data Availability Statement: The database generated and /or analysed during the current study are not publicly available due to privacy, but are available from the corresponding author on reasonable request.
Author Contributions: Conceptualization was carried out by KD; Methodlogy, YG Formal analysis, YG Data Curation, YG Wrote original draft, YG, AKV; Review and editing, KD, YG, AKV; Supervision, KD.
Funding Information: No Applicable.
REFERENCES
Adeghate, E., Schattner, P., and Dunn, E. (2006). An update on the etiology and epidemiology of diabetes mellitus. Annals of the New York Academy of Sciences, 1084, 1–29. https://doi.org/10.1196/annals.1372.029
Alam, S., Hasan, M. K., Neaz, S., et al. (2021). Diabetes Mellitus: Insights from Epidemiology, Biochemistry, Risk Factors, Diagnosis, Complications and Comprehensive Management. Diabetology, 2(2), 36–50. https://doi.org/10.3390/diabetology2020004
Alharbi, K. K., Ali Khan, I., Syed, R., et al. (2015). Association of JAZF1 and TSPAN8/LGR5 variants in relation to type 2 diabetes mellitus in a Saudi population. Diabetology & Metabolic Syndrome, 7(1), 92. https://doi.org/10.1186/s13098-015-0091-7
Anjana, R. M., Pradeepa, R., Deepa, M., et al. (2011). Prevalence of diabetes and prediabetes (impaired fasting glucose and/or impaired glucose tolerance) in urban and rural India: Phase I results of the Indian Council of Medical Research-INdia DIABetes (ICMR-INDIAB) study. Diabetologia, 54(12), 3022–3027. https://doi.org/10.1007/s00125-011-2291-5
Basile, K. J., Johnson, M. E., Xia, Q., et al. (2014). Genetic susceptibility to type 2 diabetes and obesity: Follow-up of findings from genome-wide association studies. International Journal of Endocrinology, 2014, 769671. https://doi.org/10.1155/2014/769671
Chen G., Xu Y., Lin Y., et al. (2013). Association study of genetic variants of 17 diabetes-related genes/loci and cardiovascular risk and diabetic nephropathy in the Chinese She population. Journal of Diabetes, 5(2), 136–145. https://doi.org/10.1111/1753-0407.12025
Chou, S. H.Y., Shulman, J. M., et al. (2013). Genetic Susceptibility for Ischemic Infarction and Arteriolosclerosis Based on Neuropathologic Evaluations. Cerebrovascular Diseases, 36(3), 181–188. https://doi.org/10.1159/000352054
Deepa, M., Grace, M., Binukumar, B., et al. (2015). High burden of prediabetes and diabetes in three large cities in South Asia: The Center for cardio-metabolic Risk Reduction in South Asia (CARRS) Study. Diabetes Research and Clinical Practice, 110(2), 172–182. https://doi.org/10.1016/j.diabres.2015.09.005
Fogarty, M. P., Panhuis, T. M., Vadlamudi, S., et al. (2013). Allele-specific transcriptional activity at type 2 diabetes-associated single nucleotide polymorphisms in regions of pancreatic islet open chromatin at the JAZF1 locus. Diabetes, 62(5), 1756–1762. https://doi.org/10.2337/db12-0972
Geng, T., and Huang, T. (2020). Gene-environment interactions and type 2 diabetes. Asia Pacific Journal of Clinical Nutrition, 29(2), 220–226. https://doi.org/10.6133/apjcn.202007_29(2).0002
Gong, Y., Luo, L., Wang, L., et al. (2021). Association of MTHFR and ABCB1 polymorphisms with MTX-induced mucositis in Chinese paediatric patients with acute lymphoblastic leukaemia, lymphoma or osteosarcoma—A retrospective cohort study. Journal of Clinical Pharmacy and Therapeutics, 46(6), 1557–1563. https://doi.org/10.1111/jcpt.13505
Grarup, N., Andersen, G., Krarup, N. T., et al. (2008). Association Testing of Novel Type 2 Diabetes Risk Alleles in the JAZF1, CDC123/CAMK1D, TSPAN8, THADA, ADAMTS9, and NOTCH2 Loci With Insulin Release, Insulin Sensitivity, and Obesity in a Population-Based Sample of 4,516 Glucose-Tolerant Middle-Aged Danes. Diabetes, 57(9), 2534–2540. https://doi.org/10.2337/db08-0436
Han, X., Luo, Y., Ren, Q., et al. (2010). Implication of genetic variants near SLC30A8, HHEX, CDKAL1, CDKN2A/B, IGF2BP2, FTO, TCF2, KCNQ1, and WFS1 in Type 2 Diabetes in a Chinese population. BMC Medical Genetics, 11(1), 81. https://doi.org/10.1186/1471-2350-11-81
IDF Diabetes Atlas, 9th edn. Brussels, & Belgium. (2019). IDF Diabetes Atlas, 9th edn. https://www.diabetesatlas.org
Koontz, J. I., Soreng, A. L., Nucci, M., et al. (2001). Frequent fusion of the JAZF1 and JJAZ1 genes in endometrial stromal tumors. Proceedings of the National Academy of Sciences of the United States of America, 98(11), 6348–6353. https://doi.org/10.1073/pnas.101132598
Li, J.W., Lee, H.M., Wang, Y., et al. (2016). Interactome-transcriptome analysis discovers signatures complementary to GWAS Loci of Type 2 Diabetes. Scientific Reports, 6(1), 35228. https://doi.org/10.1038/srep35228
Mahajan, A., Go, M. J., Zhang, W., et al. (2014). Genome-wide trans-ancestry meta-analysis provides insight into the genetic architecture of type 2 diabetes susceptibility. Nature Genetics, 46(3), 234–244. https://doi.org/10.1038/ng.2897
Misra, A., and Khurana, L. (2011). Obesity-related non-communicable diseases: South Asians vs White Caucasians. International Journal of Obesity (2005), 35(2), 167–187. https://doi.org/10.1038/ijo.2010.135
Morris, A. P. (2018). Progress in defining the genetic contribution to type 2 diabetes susceptibility. Current Opinion in Genetics & Development, 50, 41–51. https://doi.org/10.1016/j.gde.2018.02.003
Nakajima, T., Fujino, S., Nakanishi, G., et al. (2004). TIP27: A novel repressor of the nuclear orphan receptor TAK1/TR4. Nucleic Acids Research, 32(14), 4194–4204. https://doi.org/10.1093/nar/gkh741
Park, J. H., Ha, K. H., Kim, B. Y., et al. (2020). Trends in Cardiovascular Complications and Mortality among Patients with Diabetes in South Korea. Diabetes & Metabolism Journal, 45(1), 120–124. https://doi.org/10.4093/dmj.2020.0175
Peng, D., Wang, J., Zhang, R., et al. (2017). CDKAL1 rs7756992 is associated with diabetic retinopathy in a Chinese population with type 2 diabetes. Scientific Reports, 7(1), 8812. https://doi.org/10.1038/s41598-017-09010-w
Schulze, M. B., Rimm, E. B., Li, T., et al. (2004). C-Reactive Protein and Incident Cardiovascular Events Among Men With Diabetes. Diabetes Care, 27(4), 889–894. https://doi.org/10.2337/diacare.27.4.889
Soltanian, A. R., Hosseini, B., Mahjub, H., et al. (2020). A Bayesian analysis for investigating the association between rs13266634 polymorphism in SLC30A8 gene and type 2 diabetes. Journal of Diabetes & Metabolic Disorders, 19(1), 337–342. https://doi.org/10.1007/s40200-020-00514-3
Song, M., Zhao, F., Ran, L., et al. (2015). The Uyghur Population and Genetic Susceptibility to Type 2 Diabetes: Potential Role for Variants in CDKAL1, JAZF1, and IGF1 Genes. OMICS: A Journal of Integrative Biology, 19(4), 230–237. https://doi.org/10.1089/omi.2014.0162
Stuebe, A. M., Wise, A., Nguyen, T., et al. (2014). Maternal Genotype and Gestational Diabetes. American Journal of Perinatology, 31(1), 69–76. https://doi.org/10.1055/s-0033-1334451
Suzuki, K., Akiyama, M., Ishigaki, K., et al. (2019). Identification of 28 new susceptibility loci for type 2 diabetes in the Japanese population. Nature Genetics, 51(3), 379. https://doi.org/10.1038/s41588-018-0332-4
Yang, H., He, J., Xu, X. L., et al. (2015). Molecular characterization and tissue expression profile analysis of the porcine JAZF1 gene. Genetics and Molecular Research: GMR, 14(1), 542–551. https://doi.org/10.4238/2015.January.26.9
Zano, S., Rubab, Z. E., Baig, S., et al. (2020). Association of the JAZF1 Variant in Adults With a Parental History of Type 2 Diabetes Mellitus In Pakistan. Cureus, 12(12), e11930. https://doi.org/10.7759/cureus.11930
Zhang, Y., Santosa, A., Wang, N., et al. (2019). Prevalence and the Association of Body Mass Index and Other Risk Factors with Prediabetes and Type 2 Diabetes Among 50,867 Adults in China and Sweden: A Cross-Sectional Study. Diabetes Therapy, 10(6), 2061–2077. https://doi.org/10.1007/s13300-019-00690-3
Zhu, Y., Liu, Q., Zhou, Z., et al. (2017). PDX1, Neurogenin-3, and MAFA: Critical transcription regulators for beta cell development and regeneration. Stem Cell Research & Therapy, 8(1), 240. https://doi.org/10.1186/s13287-017-0694-z
Zimmet, P. Z. (2017). Diabetes and its drivers: The largest epidemic in human history? Clinical Diabetes and Endocrinology, 3(1), 1. https://doi.org/10.1186/s40842-016-0039-3


