1Department of Microbiology and Biotechnology, Gujarat University School of Sciences, Ahmedabad, Gujarat, India.
2L.J. School of Applied Sciences L.J.U., L.J. Campus, S.G. Road, Ahmedabad, Gujarat, India.
3Xavier’s Research Foundation, St. Xavier College Campus, Ahmedabad, Gujarat, India.
Corresponding author email: viral3007@gmail.com
Article Publishing History
Received: 14/07/2022
Accepted After Revision: 24/09/2022
Haloalkaliphilic bacteria are a specific group of bacteria known to us. The diversity of microorganisms is critical to the functioning of the ecosystem as there is the need to maintain ecological process such as decomposition of organic matter, nutrient cycling, soil aggregation and control of pathogens within the ecosystem. Microbial diversity as an indicator of the quality of agroecosystems has been widely debated, In the present study various saline soil samples collected from Bhavnagar and Uncha Kotda, Gujarat, India. The collected five samples were analysed for diversity study of soil sample for physicochemical analysis like pH, redox potential, conductivity, humidity, salinity and soil analysis for total nitrogen and organic carbon analysis also.
Total 55 haloalkaliphilic bacterial morphotypes were isolated and screened for alkaline protease production on halophilic agar medium. Out of them 76.4% gram positive bacilli, 21.8% gram positive cocci and 0.0002% were gram negative short rods. From total 55 morphotypes 33% were different morphotypes, 8% were zone of casein producer, and 16% of them were pigment producing morphotypes. All the pigmented colony producer morphotypes showed growth of orange, yellow, red, light pink and light-yellow colonies on 15%, 20%, 25%, 30% NaCl containing medium. Dominantly bacilli were found in all five samples.
Diversity indices for metabolic characterization studies like Shannon-weiner index (H’), Richness, Evenness, Cho-1, Simpson’s index and Good’s coverage were calculated based on the site wise obtained different morphotypes. Phenotypic characteristics were studied. The secondary screening and tertiary screening were done on the basis of REA and different NaCl concentration accordingly. Identification of all haloalkaliphilic protease producers were confirmed by 16S r-RNA identification.
Correlation Analysis, Dandograph, K-Means Cluster Analysis, Scatter Plot, Stacked Bar Chart.
Varia A. D, Tipre D. R, Dave S. R, Shukla V. Y. On the Diversity and Biogeography of Haloalkaliphilic Bacterial Communities Producing Alkaliphilic Protease. Biosc.Biotech.Res.Comm. 2022;15(3).
Varia A.D, Tipre D. R, Dave S. R, Shukla V. Y. On the Diversity and Biogeography of Haloalkaliphilic Bacterial Communities Producing Alkaliphilic Protease. Biosc.Biotech.Res.Comm. 2022;15(3). Available from: <a href=”https://bit.ly/3B0ijOq“>https://bit.ly/3B0ijOq</a>
Copyright © This is an Open Access Article distributed under the Terms of the Creative Commons Attribution License (CC-BY). https://creativecommons.org/licenses/by/4.0/, which permits unrestricted use distribution and reproduction in any medium, provided the original author and sources are credited.
INTRODUCTION
Haloalkaliphilic organisms are essential for fundamental research and biotechnology perspectives (Ouelhadj et al. 2020). Extremophilic organisms have unique adaptation strategies that give them an integral role in the remediation of polluted sites. Haloalkaliphilic peptide degrading bacteria is one of those groups. Higher salt concentration is necessary for growth of haloalkaliphiles for optimum growth, which is making those morphotypes and their products more suitable for use in a variety of all pharmaceutical sectors (Mechri et al. 2019; Rathakrishnan and Gopalan 2022).
Haloalkaliphilic organisms have adapted physiological mechanisms to survive with high pH and salinity in these extreme environments. Haloalkaliphilic bacterial cells cope with the high salinity of the environment and compensate to prevent osmotic stress and water leakage. These organisms synthesise some osmoregulators of organic and inorganic compounds that avoid water loss. On the basis of intracellular accumulation of inorganic ions K+ and Cl–, halophilic archaea contains the salt in strategy which provide an osmotic balance (Mainka et al. 2021). Haloalkaliphiles are present in archaea, bacteria and eukarya all the three domains of life. Based on level of salt tolerance halophiles are classified as halotolerant, slight, moderate and extreme halophilic microorganisms. Halotolerant can grow, in saline environmenta but do not always required higher salt concentration for growth (Sysoev et al. 2021; Rathakrishnan and Gopalan 2022).
True halophiles are classified as slight (1-3% NaCl), moderate (3-15% NaCl) and extreme halophiles (15-30% NaCl) with comparision to sea water salinity approximately 3.2% according to the salt concentration they require for the growth (Sysoev et al. 2021). Microbial biodiversity is the degree of variation of life forms within a biome, ecosystem and biome that measures the health of an ecosystem. Microbes maintain life over areas with extreme physicochemical conditions like hot springs, acidic springs, saline-alkaline lakes, hot and cold deserts, ocean beds that are harsh for life (Delgado-García et al. 2019). The enzymes isolated from saline bacteria have unique characteristics compared to non-halophilic bacterial enzymes (Rathakrishnan and Gopalan 2022).
Habitats of hypersaline environments are extreme with limited microbial diversity because of the combined effects of several environmental factors, including high salt concentrations, temperature, pH, low nutrient and oxygen availability(Ahmed and Mishra 2022). Biodiversity is a multidimensional property of a natural system (Santl-Temkiv et. al. 2022). Shannon and Simpson diversity indices include the measurement of variety and community heterogeneity. A Shannon index measuring the object of information theory is the content system order or disorder; frequently, it describes individual species’ condition and uncertainty. The higher the uncertainty, the higher is the diversity that depends on two factors like the species richness and the evenness of species individual distribution. A large number of species can increase diversity (Davies et. al. 2022).
Diversity can uniformly increase with the uniform distribution of species among the different sites. If each individual belongs to another species, the diversity index is the largest, and if it belongs to the same species, its diversity index is the smallest. Diversity index is a quantitative measure that reflects how many different species in the community can simultaneously take into account the phylogenetic relations among the individuals distributed among those types (Davies et. al. 2022).
Simpson diversity index ‘D’ prospect that two randomly sampled individuals belong to different species. The more significant number of species in the community is the more uniform distribution of various individuals (Thumar and Singh 2009; Mahmood et al. 2022). The higher the index indicates a good diversity of the community. Simpson index is a scalar of α-diversity. The greater the Simpson index, the higher the diversity. Rare species play a minor role in this index, while common species play a more significant role. Simpson index is more weighted on dominant species than Shannon index (Mahmood et al. 2022). This study reports the cultivable bacteria diversity with particular reference to alkaline protease producers of saline soil from various samples collected from Bhavnagar and Uncha Kotda, Gujarat, India.
MATERIAL AND METHODS
Five different saline soil samples were collected from various locations of the coastal area of Bhavnagar (21.7645 “N to 72.1519 “E) and Uncha Kotda (21.1274 “N to 71.9705 “E), Gujarat, India. Distance between the two points of the sample collection was about 30 to 50 m. Samples were collected in sterile zip lock pouches. Before analysing soil samples, all the soil samples were suspended in the distilled water (1:4 w/v) separately and allowed to settle the particles (Chaudhari 2013). Physico-chemical analysis like pH was measured using a glass-calomel combined electrode (HM digital pH meter, India) from the sample suspensions, and humidity was measured using a portable analyser (Infrared thermometer, India). The soil sample conductivity was analysed by a portable multi-meter analyser (Aquasol Power Max, India). Total nitrogen and organic carbon were measured accordingly by the Kjeldahl method and Walkley-Black chromic acid wet oxidation manual method (Vera-Gargallo and Ventosa 2018). Salinity was measured by a meter (Lutron salt meter PSA311, India).
For isolation all the five collected samples were separately added in sterile distilled water to prepare 10% w/v solutions, which were serially diluted up to 10-3 using sterile distilled water in triplicate. After serial dilutions, all the samples were plated on (i) haloalkaliphilic agar medium (Hi-media, India) supplemented with casein 5% (ii) Skimmed milk agar medium (iii) Alkaliphilic agar medium and Nutrient agar medium. All the media were supplemented with 10% NaCl and pH was adjusted to 10.5±0.5 using 1 N NaOH. After inoculation, all the plates were incubated at room temperature (34±4ºC) for 48-72 h.
During the incubation period at regular intervals, colonies showing diverse visible morphological characteristics were picked up and transferred in the respective medium sequentially three to four times to get pure isolated culture. All the pure morphotypes were preserved at 4ºC on the individual medium up to analysis (Vijayaraghavan et al. 2012; Vaishnav et al. 2014; Jothi et al. 2015; Maruthiah et al. 2015; Kim et al. 2017).In primary screening all the morphotypes were characterised based on the different morphotypes of colony characters, colonies with and without zone of casein hydrolysis, cell morphology, site-wise different types, total count and pigmented colonies. Moreover, the morphotypes were selected based on their growth in 10% NaCl and pH 10.5±0.5 containing alkaline media.
Alpha diversity marks concise the form of an ecological community concerning its richness and evenness as many contents affect the alpha diversity of a group summarising and comparing community structure by alpha diversity. In microbial ecology, analysing the alpha diversity of primer sequencing data is a common first approach to assess differences between environments. Based on the phenotypic characteristics of bacterial morphotypes, the Diversity indices, Shannon Weiner diversity index (H’), Richness (Rmargalef, Rmenhinik) and Evenness (EPielou), Chao-1 were calculated using the standard formula (Oueriaghli et al. 2014).
Simpson’s index (D) was also calculated for diversity, reciprocal and evenness. Good’s Coverage was used for checking the percent of total species present in the samples (Martínez-Olivas et al. 2019; Sorokin et al. 2022). All the above-mentioned diversity indices were calculated using ‘PAST’ software (PAST 4.03, Paleontological Statistics). Diversity indices were calculated on the basis of results of primary screening. Biochemical tests like the presence of endospore and capsule, production of
catalase, oxidase, gelatinase, amylase and lipase, vancomycin test and 3% KOH test for Gram reaction were performed for all selected bacterial morphotypes (Chen et al. 2022). All the selected morphotypes were used to check relative enzyme activity (REA) and colony characters in secondary screening on 10% to 25% NaCl containing a haloalkaliphilic medium (Gaffney et. al. 2021). The morphotypes were selected for the tertiary screening based on the high REA results on different NaCl containing medium plates (Ariaeenejad et al. 2022).
The morphotypes from secondary screening were selected for final tertiary screening based on their growth in different NaCl containing broth and REA. In tertiary screening, protease production was checked in a production broth medium containing NaCl and alkaline pH. The salt concentrations studied was from 5% to 25%, while the medium pH tested were 9, 10, 11 at each NaCl concentration in the production medium. The composition of haloalkaliphilic protease production medium consist (g/L): yeast extract, 1.0; glucose, 6.0; malt extract, 1.0; FeSO4.7H2O, 0.1; MgSO4.7H2O, 0.5; K2HPO4, 0.5; peptone, 2; NaCl, 10.0; casein, 1.0 (Gupta et al. 2015). All the selected morphotypes were categorised as slight halophile or halotolerant (1-5% or 0.2-0.85 M NaCl), moderate halophiles (5-20 % or 0.85-3.4 M NaCl) and extreme halophiles (20-30 % or 3.4-5.1 M NaCl) (Bhatt and Singh 2017; Upadhyay et al. 2019; Asitok et al. 2022).
Species identification of morphotype were confirmed by 16S rRNA gene sequencing. The 16S rRNA gene sequences of selected all morphotypes were also submitted to the GenBank and sequence id accession number were obtained. All the obtained sequences were aligned with a multiple sequence alignment and phylogenetic tree of the morphotypes was constructed by the neighbour joining analysis using “Molecular Evolutionary Genetics Analysis Version 7.0 software (Saitou and Nei 1987; Kumar et al. 2016; Zuo et al. 2022).
RESULTS AND DISCUSSION
Physico-chemical characters of collected soil samples: The results of the physicochemical analysis of all the samples are presented in Table 1. The pH of the samples collected from Bhavnagar ranged from 10.5 to 11.62, and samples from Uncha Kotda ranged from 11.12 to 11.73. The alkalinity of the samples was due to the presence of different salts present in the saline soil. The temperature at the time of sample collection was 34±5ºC. The redox potential of the samples collected from Bhavnagar ranged from 105 to 122 mV, and samples from Uncha Kotda ranged from 130 to 150 mV. The conductivity ranged from 15.69 to 36.9 mS and 13.08 to 18.7 mS for samples from Bhavnagar and Uncha Kotda, respectively.
Table 1. Physico-chemical analysis of the samples
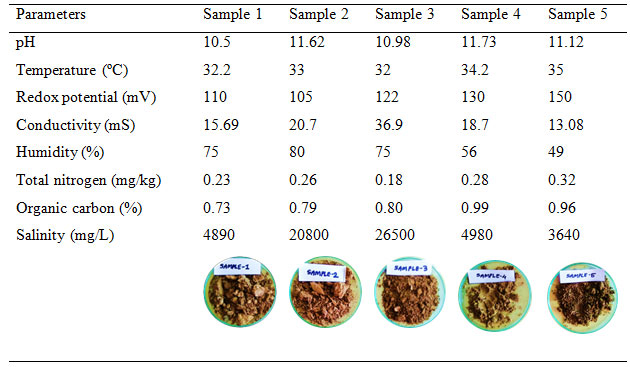
Total nitrogen and organic carbon ranged from 0.18 to 0.32 mg/kg and 0.73 to 0.96% respectively. While salinity of the samples from Bhavnagar ranged from 4890 to 26800 mg/L, and samples from Uncha Kotda ranged from 3640 to 4980 mg/L. Salinity and conductivity have a linear relationship. Results here mentioned in table 1. As salinity increases, the conductivity of the sample also increases (Rusydi, 2018). Samples (1, 2 and 3) from Bhavnagar showed more redox potential, salinity, and conductivity than samples (4 and 5) from Uncha Kotda. The results measured here are of one specific day of sample collection, and hence the variation was due to the different sample collection sites.
The collected saline soil samples were diverse in physicochemical characters and therefore showed variation in the results from site to site. The results indicated the presence of dissolved solids, salts and impurities in the form of hydroxides, carbonates and bicarbonates with sodium chloride imparting a slightly alkaline nature to the habitat (Dave and Desai, 2006). Moreover, Bhavnagar and Uncha Kotda are situated 84 km away from each other.
Isolation of haloalkaliphilic morphotypes: The haloalkaliphilic morphotypes were isolated from saline soil of Bhavnagar and Uncha Kotda, Gujarat, India. Total 55 morphologically different types (morphotypes) of bacteria were isolated on a 10% NaCl containing medium. Of the obtained bacterial morphotypes, 9.18% were halotolerant, 9.6% were moderate halophiles, and 11% were extreme halophiles. Table 2 shows the site-wise total count of obtained bacterial morphotypes.
Table 2. Site-wise total bacterial count on various media
| Medium | Total viable count (CFU/ g) | ||||
| Sample number | |||||
| 1 | 2 | 3 | 4 | 5 | |
| Haloalkaliphilic medium
(Jothi et al. 2015) |
5.65×104 | 8.01×104 | 9.06×104 | 2.36×104 | 2.89×104 |
| Alkaliphilic medium (Vijayaraghavan et al. 2012) | 0.68×103 | 0.044×103 | 0.88×102 | 0.03×102
|
0.5×102 |
| Skimmed milk agar medium (Vaishnav et al. 2014) | 0.12×101 | 0.03×101 | 0.05×101
|
0.4×102 | 0.02×102 |
| Nutrient agar medium | 0.7×101 | 0.5×101 | 0.3×101 | 0.8×101 | 0.01×101 |
Saline soil samples collected from Bhavnagar showed more morphotypes as compared to Uncha Kotda. The majority of the morphotypes were found present in each sample. Table 2 shown the site-wise bacterial count results obtained on the studied different media, and the haloalkaliphilic medium containing 10% NaCl and 5% casein was the most suitable one. Site 3 showed the highest 9.06×104 CFU/g. In contrast, skimmed milk agar and nutrient agar medium resulted in the lowest bacterial count. Haloalkaliphilic agar medium was selected as an appropriate medium for the isolation and further screening of the bacteria from the saline soil.
Primary screening: Colony and cellular characters were recorded in terms of all the colony characters like size, shape, margin, elevation, texture, opacity and pigmentation as mentioned in Table 3, with particular prominence on pigment colour and casein degradation zone. Among 55 morphotypes, cellular characteristics showed the dominance of 39 gram-positive bacilli, and the remaining were 15 gram-positive cocci and one gram-negative short rod. The obtained morphotypes showed growth on 10, 15 and 20% NaCl concentrations. All the samples showed the presence of a wide variety of haloalkaliphilic protease producing bacteria. A total of 40 morphotypes showed a zone of casein hydrolysis that were screened out after primary screening. Morphologically different morphotypes with a zone of casein hydrolysis were used for secondary screening.
Site-3 gave the highest CFU/g (Table 2) of the collected soil sample. Site-1 gave the highest number of bacterial morphotypes. It was also observed that sample 1 showed the maximum bacterial diversity at alkaline pH and higher NaCl concentration. The colony characters of 55 morphotypes are mentioned in Table 3. Many morphotypes were common to all sites. Out of these 55 morphotypes, 41 were gram-positive bacilli comprising 76.4%, 13 gram-positive cocci (21.8%), and only one isolate (0.0002%) gram-negative short rod (Qu et al. 2022).
Site-wise total morphotypes, morphotypes with a zone of casein hydrolysis and their morphology are mentioned in Figure 1 of correlation analysis (Minitab software). Figure 1 depicts that the correlation coefficient ranges from -0.94 to 1.0. We found maximum morphotypes of bacilli as compared to cocci and short rods, respectively. Bacilli to cocci correlation observed were -0.94 and -0.09 in the case of bacilli to short rods. Total morphotypes found were between -0.82 to 1.0, bacilli with 0.80, cocci with -0.82 and short rod with -0.04.
This analysis suggests the diversity of species similarity of all sites. From total morphotypes, casein hydrolysing zone forming morphotypes were in the range of 0.19 to 1.0. Halophilic morphotypes are pleomorphic as they can survive in high NaCl concentrations (Patel et al. 2006; Yadav and Patil 2020).Halobacteriales needs high salt concentration for their structural steadiness. The non-coccoid forms of bacterial structure lie under 10% NaCl concentration (Dave and Desai 2006). Here majority of morphotypes were gram-positive bacillus (Qu et al. 2022).
Figure 1: Correlation analysis of 55 morphotypes based on the zone of casein hydrolysis and their morphology
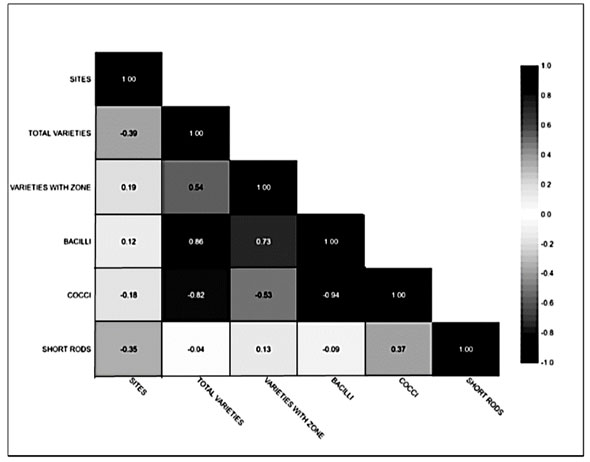
This statistical (Minitab 20) technique was used to determine the dependence between two or more variables. Here this correlation design shown a positive correlation ship between all variables. It offers a strong dependency relation of variables pairs. The extent to which two variables vary together were also determined apart from the relationship of strength and direction.
Site wise study of pigmented morphotypes: Out of 55 morphotypes, 33% were different morphotypes, and from all of them, 8% of morphotypes showed a zone of casein hydrolysis, and 16% of the morphotypes produced a variety of pigments. Among the 55 morphotypes and 40 different significant casein hydrolysis zone-producing morphotypes that could grow at 10%, NaCl concentrations were selected further. Colonies of the selected 55 morphotypes showed various pigment formations like orange, red, pink, light pink, yellow on Haloalkaliphilic agar medium plates. Figure 2 represents the comparison of total bacterial morphotypes with pigmented and non-pigmented bacterial morphotypes. Sample from site 3 showed the highest number of total bacterial morphotypes with pigmented colonies. Site 2 and site 4 equally showed the highest number of pigmented colonies than the other sites (Qu et al. 2022).
Table 3. Colony characters observed on casein agar plates (pH 10.5) from samples
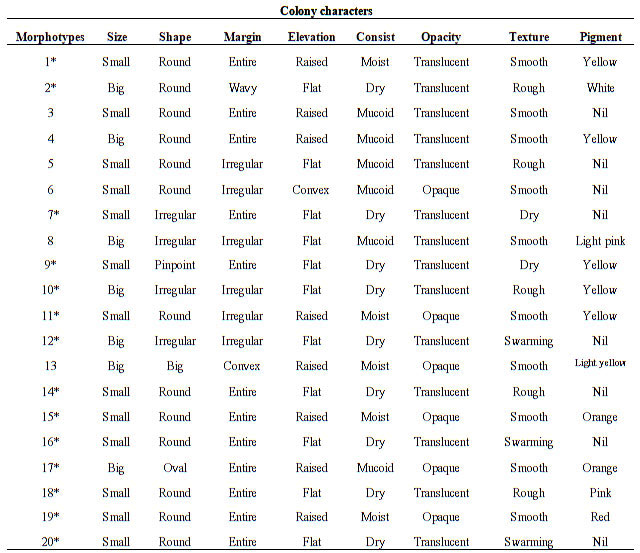
Note- ‘*’ for colonies with a zone of casein hydrolysis.
Light pink-coloured bacterial morphotypes were dominating among all the morphotypes. All the pigmented bacterial morphotypes were gram-positive bacilli or cocci. Generally dominant red and orange pigmented colonies presented at 25% NaCl, pink at 20%, yellow at 15%, whereas colourless colonies dominated at 10% NaCl concentration. The pigment intensity and pigment producer morphotypes increased with NaCl concentration in the medium (Dave and Desai 2006; Purohit et al. 2016; Qu et al. 2022).
Figure 2: Site-wise morphotypes of pigmented bacteria compared with the
total number of isolate and non-pigmented morphotypes.
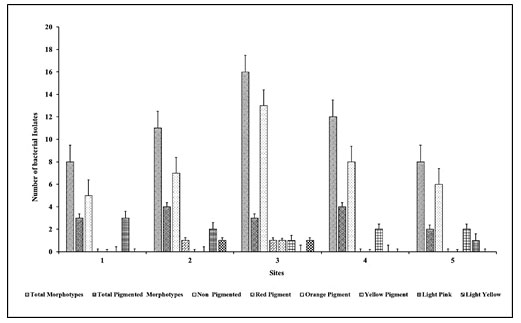
Here we can conclude that in all the collected saline soil samples, greater diversity in the bacterial colony observed on haloalkaliphilic agar, indicating that obtained morphotypes could be halophilic (Hegazy et al. 2020).
Study of diversity indices: The results of various diversity indices are shown in Figure 3. Shannon Weiner index (H’) ranged from 2.9 to 4.39. Shannon’s diversity index for bacterial community reported from 2.97 to 5.48 for microbial diversity (Maron et al. 2018). Shannon’s index was also reported 1.47 to 2.04 for Halomonas community from saline soil of Rambla Salada (Oueriaghli et al. 2014). Samples 1 and 5 had a lower value of Shannon Index above 2, whereas samples 2, 3 and 5 had 3.24, 4.39 and 3.89 accordingly. Shannon’s evenness index reported as 2.3 (Sharma et al. 2021) whereas, our results showed evenness that ranged from 1.26 to 2.62. Site 3 offered the highest evenness of 2.62. Simpson’s evenness for haloalkaliphilic archaea reported from 0.40 to 0.78 from marine samples (Martínez-Olivas et al. 2019; Hegazy et al. 2020).
Figure 3: Site-wise diversity indices calculated based on species observations
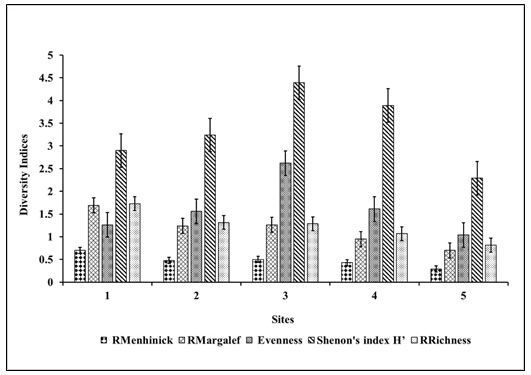
Richness ranged from 0.8 to 1.73, RMargalef ranged from 0.7 to 1.69, and RMenhinick ranged from 0.291 to 0.7. Bacterial diversity richness reported from 313 to 1004 (Maron et al. 2018). For lignite mine bacteria, RMargalef reported from 0.0 to 15.88, and RMenhinick reported from 0.87 to 5.89 (Patel et al. 2009; Maron et al. 2018). In our results, site 1 showed the highest species richness, whereas site 5 showed the lowest species richness than other sample collected sites. RMargalef and RMenhinick of 2.3 and 1.7 for haloalkaliphilic actinobacteria had been reported (Sharma et al. 2021). Relative frequency and relative density depicted in Figure 3 concerning obtained total morphotypes from all 5 sites (Sharma et al. 2021).
Variety no. 12 (Table 3) showed the highest relative frequency among all sites in the study. Site 1 gave relative density ranged from 3.8 to 23.07, whereas sites 2, 3, 4 and 5 showed relative density ranged from 8.42 to 16.34, 7.3 to 19.57, 9.25 to 21.29 12.59 to 24.44, respectively. Isotype 12 showed the highest relative frequency of presence to all the respective 5 sites. Colony types 1-10 and 12 were observed in sample 1 from site-1. Colony types 1-3 and 11-15 were present in sample 2 from site-2. Colony types 2, 8, 10-15 were observed in sample no. 3 from site-3. Colony types 12, 16-20 were present in sample no. 4 from site-4. Colony types 12, 16, 18 and 20 were present in sample no. 5 from site-5. Colony numbers 1-4 and 11 showed similar colony morphology visually that were present in samples 1 and 2 (Sharma et al. 2021; Li et al. 2022).
Figure 4: Study of relative frequency and relative density of totally different morphotypes
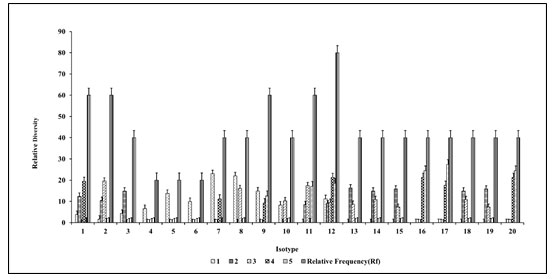
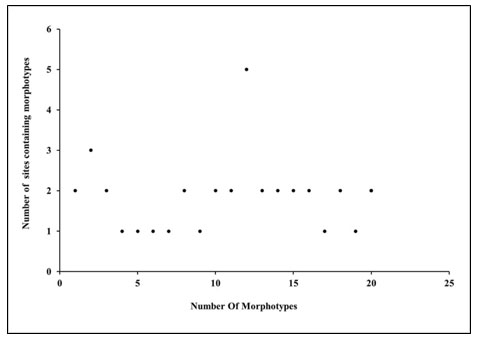
Colony numbers 2 and 10-15 were morphologically similar colonies in samples 2 and 3. Colony no. 12, 16-20 were present in samples 4 and 5. Morphotype 12 was dominantly present in all the sites. Figure 5 scatter plot showen the site-wise presence of different morphotypes based on colony morphology.
Figure 5: Site wise morphotypes (based on colony characters)
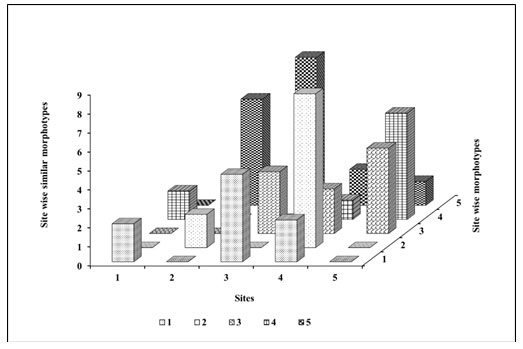
The similarity index (Figure 6) could be explained based on the ecosystem of the respective site. Site 1 showed the highest richness and diversity indices, while site 3 showed the least richness and diversity of bacterial morphotypes. All the other indices calculated were in support of these observations. Chao1 abundance-based species richness estimator for missing morphotypes was 1.3 from species morphotypes. From the study of all the diversity indices, it has been determined that sample number 1 had overall low density, whereas sample number 3 showed high density, evenness and richness.
Figure 6: Site wise similarity index
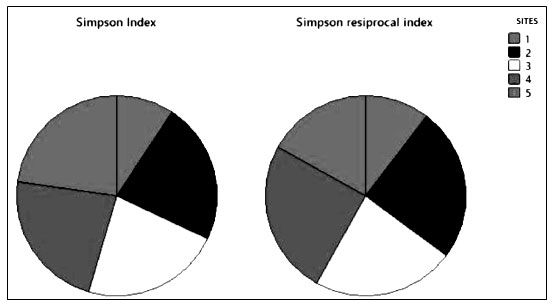
Simpson’s index (D) is shown in Figure 7. It was demonstrated the numerical proportion of similarity probability between various randomly selected species. It ranges from 0 to 1 with representing infinite diversity and no diversity, respectively. The higher the value of Simpson’s diversity index greater the ecological diversity. The ecological values of Simpson’s index site-wise were 0.4, 0.98, 0.97,0.98 and 0.98 accordingly. The Simpson’s index of diversity 0.78 shown rich haloalkaliphilic bacterial diversity. Simpson’s index has been reported for haloalkaliphilic archaeal microbial diversity from 0.70 to 0.97 and evenness from 0.07 to 0.48 (Martínez-Olivas et al. 2019). Simpson’s reciprocal index site-wise from site 1 to 5 were 25.08, 59.7, 55.4, 60.24 and 40.81 accordingly. For haloalkaliphilic bacterial archaea. It has been reported from 27.08 to 61.27 (Martínez-Olivas et al. 2019; Sharma et al. 2021).
Figure 7: Simpson’s diversity index-based comparison between the selected sites
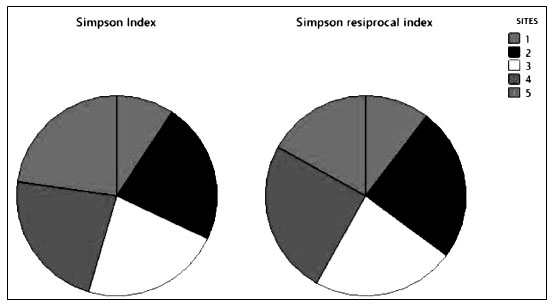
From the Shannon’s and Simpson’s indices studies, it was observed that most diverse bacterial morphotypes were able to grow at different salt concentrations with a variety of pigment production could be the reason for its high diversity value (Li et al. 2022). There was a need to evaluate how well a sample reflects the true diversity of a specific niche.
Figure 8: Site-wise Chao 1 analysis
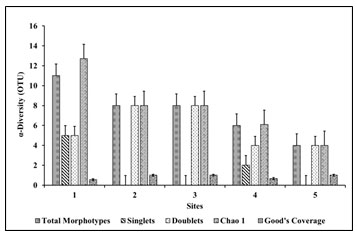
The diversity of a particular niche always synonymouses with species richness and relative abundance in time and space. Accurate assessment of species richness is instrumental for each biological community. With other indices, Chao1 was a nonparametric method for estimating the number of species in a community (Farheen et. al. 2022). Chao 1 was based on the concept that rare species infer most information about the missing species as the Chao 1 richness estimator gives more weight to the low abundance of the singletons and doubletons were used for estimation number of missing species (Oosterkamp et al. 2019; Sikorski et al. 2022).
Site-wise missing morphotypes (Figure 8) were ranged from 1.2 to 10.5. Site 1 was shown the highest value of Chao-1 analysis 12.7, whereas site 5 was shown a lower value of Chao 1 analysis 4. Site 1 revealed maximum variables and missing morphotypes of bacterial morphotypes. Oosterkamp et al. (2019) reported the Chao-1 index was from 410 to 1091 range for microbial community diversity. Whereas, for haloalkaliphilic archaeal diversity Chao-1 index of 70 to 349.14 was given by Martínez-Olivas et al. (2019). Where Zhang et al. (2018) reported Chao-1 index from 74.33 to 799.0 for microbial diversity.
Good’s coverage (Coverage= 1-(singlets/total morphotypes)) of all the sites 1 to 5 was calculated as, 0.37%, 0.32%, 0.67% and 0.63% respectively. It estimates the percentage of total species present in the sample. It was an alpha diversity metric. Good’s coverage was reported for bacterial diversity in the range of 0.50% to 0.82% and halophilic archaea 0.55% to 0.94% (Martínez-Olivas et al. 2019). Moreover, Zhang et al. (2018) reported Good’s Coverage value ranged from 0.38% to 2.47% for microbial diversity.
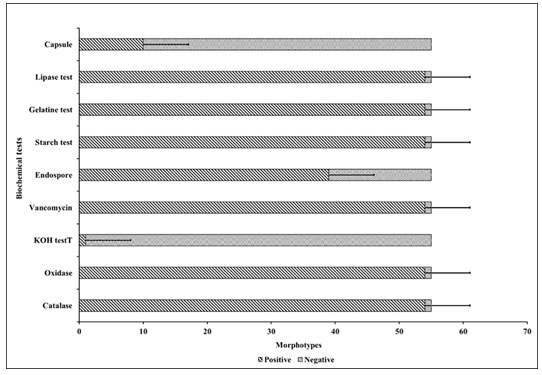
Phenotypic characterisation of organisms: Figure 9 represents the cluster bar chart for all 9 biochemical tests. This analysis showed a pictorial representation analysis of biochemical tests of 55 bacterial morphotypes obtained on the haloalkaliphilic agar medium. Clustered bar chart allows the direct comparison of multiple data series per category, which shown change over time. Each sector denotes a compatible part of the whole. The figure represented the categorical data of catalase, oxidase, KOH, vancomycin, endospore, capsule, gelatine degradation, lipase production, and starch hydrolysis tests.
Total 54 morphotypes showed catalase, oxidase, KOH, vancomycin, gelatine, lipase and starch hydrolysis test positive. At the same time, one isolate showed these mentioned tests negative. Sixteen morphotypes showed the formation of endospore, and 39 gave the test negative. Ten morphotypes showed the presence of a capsule, and 45 morphotypes were non-capsule former. It offers the inner products between the observations and variables. One bacterial isolate showed a difference in most biochemical tests in the plot that correlates to the Chol-1 index whereas, the rest of the bacterial morphotypes showed similarity. It could be due to its various metabolic properties (Sikorski et al. 2022).
Figure 9: Cluster bar chart for the biochemical test of 55 morphotypes
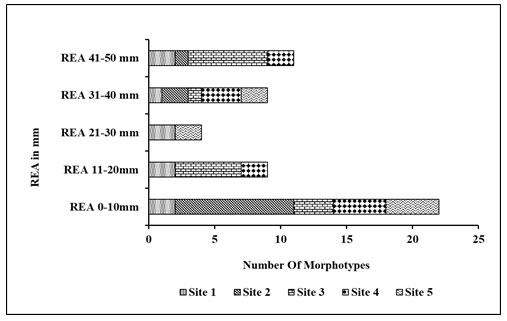
Secondary screening based on REA: After the primary screening, all the obtained 40 morphotypes spreaded on the haloalkaliphilic agar medium of pH 10.5±0.5 and supplemented with 5% skimmed milk containing different NaCl concentrations of 10% to 25% (Figure 10). Site 3 was found rich in a number of bacterial morphotypes shown zone of casein hydrolysis at 15%, 20% and 25% NaCl concentration. Therefore, 20 morphologically distinct morphotypes were selected for the salt tolerance test from 20 % NaCl and pH 10.5±0.5 containing haloalkaliphilic agar medium with higher REA activity. The relative enzyme activity (REA) of the selected 40 morphotypes shown in Figure 11. REA values of 21 to 40 mm giving 20 morphotypes were further selected for the haloalkaliphilic protease production test.
Based on REA, the morphotypes can be categorised into three groups. (REA= Zone diameter of casein hydrolysis/Colony diameter in mm). REA>5mm is magnificent, REA >2 to 5 mm is satisfactory and REA<2 is the deficient producer of protease (Jadhav et al. 2016). Here, the zone of casein hydrolysis on haloalkaliphilic agar medium shown the presence of protease producing bacteria. Some strains showed clear hydrolysis zones around the bacterial colonies on the haloalkaliphilic agar medium with 10% NaCl, which indicates a dominant amount of protease (Cui et al. 2015; Sikorski et al. 2022).
Figure 10: Secondary screening based on REA
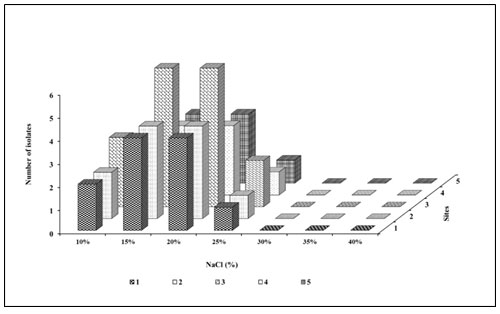
Figure 11: REA of morphotypes (Stacked bar) on the basis of zone of casein hydrolysis
Tertiary screening for haloalkaliphilic protease production with different pH and NaCl concentrations:Significant haloalkaliphilic protease producing bacteria were selected based on REA. Figure 13(a, b) shown the fermentative production of haloalkaliphilic protease.
Figure 12: REA of selected morphotypes
Figure 13: Tertiary screening of different pH and NaCl concentration (a) pH (b) NaCl
Selected 7 morphotypes denoted as 1D, 2B, 3E, 3K, 4D, 4I and 5F were checked for haloalkaliphilic protease production at different pH (9, 10 and pH 11), and NaCl concentrations (10%, 15%, 20% and 25%). At least one isolate from each site was selected. Further, the isolate that gave the best result at pH 11 and 20% NaCl concentration within 24 h will be used to optimise the enzyme production. The tertiary screening results based on pH 11 and 20% NaCl concentration, isolate 2B, 3E, 4I, 5F and 3K were selected for further experiments.
Phylogenetic analysis of the morphotypes: The biochemical tests determined the metabolic activity of the bacterial morphotypes. 16S rRNA gene sequencing of selected morphotypes were used for the identification of bacteria and phylogenic analysis as the “optimum mark”. It was reported that molecular based identification of morphotypes by 16s rRNA gene sequencing helps at the genus level identification (Patel et al. 2019). On the basis of 16S rRNA partial gene analysis selected isolates represented 24 different genera and species of that genera. By this way it was showed morphological and metabolical diversity.
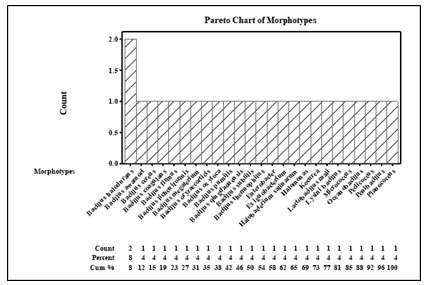
Pareto plot analysis of the morphotypes showed that most of the morphotypes belonged to the genus bacillus species and identified as Bacillus halodurans, Bacillus megaterium, Bacillus subtilis, Bacillus oxytoca, Micrococcus, Planococcus, Halomonas, Pedicoccus, Oceanobacillus spp., Exiguobacterium, Bacillus cereus, Enterobaceter aerogenes, Bacillus thermophiles, Lactobacillus mali, Bacillus licheniformis, Penibacillus, Bacillus oryzaecoeticis, Lysinibacillus, Bacillus pumilis, Bacillus coagulans, Halobacterium salinarum, Bacillus halodurans, Bacillus firmus, Bacillus awamori, Bacillus halodurans, Bacillus qingdaonensis. Bacillus halodurans was present frequently in all the sample collection sites (Sikorski et al. 2022).
Figure 14 Pareto plot for bacterial phylogenetic analysis
All the morphotypes were gram positive except one gram negative morphotype Enterobacter. Rest of the other morphotypes were present distinctively at all the sites. Based on 16S r-RNA gene analysis, evolutionary relationship between cultivable morphotypes isolated from different locations of Bhavnagar city and Uncha kotda Gujarat, India.
Figure 15: Cluster dandogram showing relationship amongst all morphotypes
Hierarchical Cluster analysis of taxonomic identification
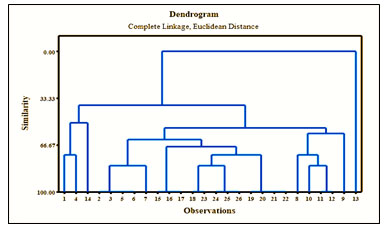
Tree diagram of taxonomic relationship for site wise morphotypes with distance and similarity shown in the figure 15. The hierarchial clustering algoritham shown similar objects into cluster groups.The end point was a set of clusters which was distinct from each other clusters and objects within each cluster are broadly similar to each other. Here, site 1, 2, 3, 4 and 1, 4 and 5 were showing diversity similarity which matches to the similarity results of the Simpson’s diversity index results mentioned above.The evolutionary distance of morphotypes is depicted by the length of the horizontal line (Daly et al. 2018).The branch point similar with the horizontal line was the divergence point of two microbial species (Mulango et al. 2020). On the basis of this diversity studies it has been concluded that the hypersaline soil sample containing all the sites were augmented with a vast number of morphotypes showing divergent metabolic potential (Sikorski et al. 2022).
CONCLUSION
The study of culture-dependent biodiversity from Bhavanagr and Uncha Kotda from Gujarat coast showed species richness and evenness. Site 3 from Bhavnagar was offering the highest variation in the diversity study. Moderately halophilic bacterial morphotypes were selected for further analysis. Haloalkaliphilic bacteria are capable of showing function under higher alkaline pH and salt conditions. They showed diversity based on their colony correlated cultural and morphological features, gram reaction, biochemical properties, secretion production of enzymes and metabolites. Diversity, physiology and metabolic studies of haloalkaliphilic bacteria help understand biopower and surveillance of these organisms under extreme conditions.
Here, we can conclude that in all the collected samples, we observed a greater number of cultivable morphotypes with different pigments on haloalkaliphilic agar that indicate isolated morphotypes to be halophilic bacteria. Further halophilic aspects of morphotypes will be confirmed by SDS-PAGE analysis by correlation of molecular weight of the isolated morphotypes with standard molecular weight of haloalkaliphiles. All selected five morphotypes that indicated a potential for high production of haloalkaliphilic protease would be used for scale-up of process application.
ACKNOWLEDGEMENTS
The study was supported by the teaching and non-teaching staff of the Department of Microbiology, L.J. IAS, Ahmedabad and L.J.K. trust.
REFERENCES
Ahmad A, and Mishra R (2022). Structural and functional adaptation in extremophilic microbial α-amylases. Biophysical Reviews, pp.1-17 (14):499-515. https://doi.org/10.1007/s12551-022-00931-z.
Ariaeenejad S, Kavousi K and Mamaghani et al., (2022). Simultaneous hydrolysis of various protein-rich industrial wastes by a naturally evolved protease from tannery wastewater microbiota. Science of The Total Environment. 815. 152796. https://doi.org/10.1016/j.scitotenv.2021.152796.
Asitok A, Ekpenyong M, Takon I et al., (2022). Overproduction of a thermo-stable halo-alkaline protease on agro-waste-based optimized medium through alternate combinatorial random mutagenesis of Stenotrophomonas acidaminiphila. Biotechnology Reports. 35. 00746. https://doi.org/10.1016/j.btre.2022.e00746.
Bhatt HB and Singh SP (2017). Phylogenetic and phenogram based diversity of haloalkaliphilic bacteria from the saline desert. Microbial Biotechnology: Technological Challenges and Developmental Trends. (1). 373-386. https://doi.org/10.1201/b19978.
Bhatt HB, Gohel SD and Singh SP (2018). Phylogeny, novel bacterial lineage and enzymatic potential of haloalkaliphilic bacteria from the saline coastal desert of Little Rann of Kutch, Gujarat, India. 3 Biotech. 8(1): 1-12. https://doi.org/10.1007/s13205-017-1075-0.
Chaudhari KG (2013). Studies of the physicochemical parameters of soil samples. Advances in Applied Science Research. 4(6): 246-248. http://www.pelagraresearchlibrary.com.
Chen Z, Yang G and Mu T et al., (2022). Rate-based model for predicting and evaluating H2S absorption in the haloalkaliphilic biological desulfurization process. Journal of Industrial and Engineering Chemistry. 110. 479-490. https://doi.org/10.1016/j.jiec.2022.03.020.
Cui H, Wang L and Yu Y (2015). Production and characterisation of alkaline protease from a high yielding and moderately halophilic strain of SD11 marine bacteria. Journal of Chemistry. Vol.2015: 1-8. https://doi.org/10.1155/2015/798304.
Daly AJ, Baetens JM and De Baets B (2018). Ecological diversity: measuring the unmeasurable. Mathematics. 6(7): 119. https://doi.org/10.3390/math6070119.
Dave SR and Desai HB (2006). Microbial diversity at marine salterns near Bhavnagar, Gujarat, India. Current Science. 90(4): 497-500. https://www.jstor.org/stable/24088939.
Davies NS, McMahon WJ, Shillito AP et al. (2022). Deep Time Biogeomorphology: the co-evolution of life and sediments. Palaios. 37(6). 219-223. https://doi.org/10.2110/palo.2022.029
Delgado-García M, Flores-Gallegos AC, Kirchmayr M et al. (2019). Bioprospection of proteases from Halobacillus andaensis for bioactive peptide production from fish muscle protein. Electronic Journal of Biotechnology. 39: 52-60. https://doi.org/10.1016/j.ejbt.2019.03.001.
Farheen KS, Reyes NJDG and Jeon MS et al., (2022). The Status of Ramsar wetlands in India: A review of ecosystem benefits, threats, and management strategies. Journal of Wetlands Research. 24(2). 123-141. https://doi.org/10.17663/JWR.2022.24.2.123.
Gaffney EM, Simoska O and Minteer SD (2021). The use of electroactive halophilic bacteria for improvements and advancements in environmental high saline biosensing. Biosensors, 11(2), 48. https://doi.org/10.3390/bios11020048.
Gupta M, Aggarwal S and Navani NK et al. (2015). Isolation and characterisation of a protease-producing novel haloalkaliphilic bacterium Halobiforma sp. strain BNMIITR from Sambhar Lake in Rajasthan, India. Annals of Microbiology. 65(2): 677-686. https://doi.org/10.1007/s13213-014-0906-z.
Hegazy GE, Abu-Serie MM and Abo-Elela GM et al., (2020). In vitro dual (anticancer and antiviral) activity of the carotenoids produced by haloalkaliphilic archaeon Natrialba sp. M6. Scientific Reports. 10(1). 1-14. https://doi.org/10.1038/s41598-020-62663-y.
Jadhav HP, Sonawane MS and Khairnar MH et al., (2020). Production of alkaline protease by rhizospheric Bacillus cereus HP_RZ17 and Paenibacillus xylanilyticus HP_RZ19. Environmental Sustainability. 3(1). 5-13. https://doi.org/10.1007/s42398-020-00096-z.
Kim BR, Shin J, Guevarra RB et al., (2017). Deciphering diversity indices for a better understanding of microbial communities. Journal of Microbiology and Biotechnology. 27(12): 2089-2093. https://doi.org/10.4014/jmb.1709.09027.
Kumar S, Stecher G and Tamura K (2016). MEGA7: Molecular Evolutionary Genetics Analysis version 7.0 for bigger datasets. Molecular Biology and Evolution. 33(7).1870-1874. https://doi.org/10.1093/molbev/msw054.
Li X, Yang M and Mu T et al., (2022). Composition and key-influencing factors of bacterial communities active in sulfur cycling of soda lake sediments. Archives of Microbiology. 204(6). 1-12. https://doi.org/10.1007/s00203-022-02925-7.
Mahmood MZ, Bibi S and Shahzad M et al., (2022). Mechanisms of microbes to combat salinity in soil by producing secondary metabolites. Arabian Journal of Geosciences. 15(1). 1-15. https://doi.org/10.1007/s12517-021-09371-7.
Mainka T, Weirathmüller D, Herwig C et al., (2021). Potential applications of halophilic microorganisms for biological treatment of industrial process brines contaminated with aromatics. Journal of Industrial Microbiology and Biotechnology. 48(1-2). kuab015. https://doi.org/10.1093/jimb/kuab015.
Maron PA, Sarr A and Kaisermann A et al., (2018). High microbial diversity promotes soil ecosystem functioning. Applied and Environmental Microbiology. 84(9). 1-13. https://doi.org/10.1128/AEM.02738-17.
Martínez-Olivas MA, Jiménez-Bueno NG and Hernández-García JA et al. (2019). Bacterial and archaeal spatial distribution and its environmental drivers in an extremely haloalkaline soil at the landscape scale. Peer Journal Life and Environment. 7: 6127 (1-28). https://doi.org/10.7717/peerj.6127.
Maruthiah T, Somanath B and Immanuel G et al. (2015). Deproteinization potential and antioxidant property of haloalkaliphilic organic solvent tolerant protease from marine Bacillus sp. APCMST-RS3 using marine shell wastes. Biotechnology Reports. 8: 124-132. https://doi.org/10.1016/j.btre.2015.10.009.
Mechri S, Bouacem K and Jabeur F et al., (2019). Purification and biochemical characterisation of a novel thermostable and halotolerant subtilisin SAPN, a serine protease from Melghiribacillus thermohalophilus Nari2A T for chitin extraction from crab and shrimp shell by-products. Extremophiles. 23(5): 529-547. https://doi.org/10.1007/s00792-019-01105-8.
Mulango E, Kasili R and Mwirichia R et al., (2020). Isolation and characterisation of haloalkaliphilic bacteria from the hot springs of Lake Magadi. African Journal of Microbiology Research. 14(7): 294-302. http://hdl.handle.net/123456789/1524.
Oosterkamp MJ, Bauer S and Ibáñez AB et al., (2019). Identification of methanogenesis and syntrophy as important microbial metabolic processes for optimal thermophilic anaerobic digestion of energy cane thin stillage. Bioresource Technology Reports. 7:100254 (1-10). https://doi.org/10.1016/j.biteb.2019.100254.
Ouelhadj A, Bouacem K and Asmani KL et al., (2020). Identification and homology modeling of a new biotechnologically compatible serine alkaline protease from moderately halotolerant Gracilibacillus boraciitolerans strain LO15. International Journal of Biological Macromolecules. 161:1456-1469. https://doi.org/10.1016/j.ijbiomac.2020.07.266.
Oueriaghli N, González-Domenech CM and Martínez-Checa F et al., (2014). Diversity and distribution of Halomonas in Rambla Salada, a hypersaline environment in the southeast of Spain. FEMS microbiology ecology. 87(2): 460-474. https://doi.org/10.1111/1574-6941.12237
Patel M, Tipre DR and Dave SR (2009). Microbial diversity by substrate utilisation profiles of lignite mines samples of Gujarat, India. In Advanced Materials Research. Trans Tech Publications Ltd.71: 101-104. https://doi.org/10.4028/www.scientific.net/AMR.71-73.101.
Patel RK, Dodia MS and Joshi RH et al., (2006). Purification and characterisation of alkaline protease from a newly isolated haloalkaliphilic Bacillus sp. Process Biochemistry. 41(9). 2002-2009. https://doi.org/10.1016/j.procbio.2006.04.016.
Patel R, Dodia M, and Singh SP (2005). Extracellular alkaline protease from a newly isolated haloalkaliphilic Bacillus sp.: Production and optimisation. Process Biochemistry. 40(11). 3569-3575. https://doi.org/10.1016/j.procbio.2005.03.049.
Purohit MK, Raval VH and Singh SP (2014). Haloalkaliphilic bacteria: molecular diversity and biotechnological applications. Geomicrobiology and Biogeochemistry. 39: 61-79. https://doi.org/10.1007/978-3-642-41837-2_4.
Purohit MS, Tipre DR and Shah MB et al., (2016). Isolation of halotolerant and halophilic bacteria and screening of their poly enzyme potential. 45(1):129-136.
Qu S, Liu L, Zhang L et al., (2022). Biodegradation of crude oil by a moderately haloalkaliphilic Acinetobacter strain. Petroleum Science and Technology. 1-15. https://doi.org/10.1080/10916466.2022.2041666.
Rathakrishnan D and Gopalan AK (2022). Isolation and characterization of halophilic isolates from Indian salterns and their screening for production of hydrolytic enzymes. Environmental Challenges. 6. 100426. https://doi.org/10.1016/j.envc.2021.100426.
Rusydi AF (2018). Correlation between conductivity and total dissolved solid in various type of water: A review. In IOP conference series: earth and environmental science (Vol. 118, No. 1, p. 012019). 10.1088/1755-1315/118/1/012019.
Saitou N and Nei M (1987). The neighbor-joining method: A new method for reconstructing phylogenetic trees. Molecular Biology and Evolution 4(4): 406-425. https://doi.org/10.1093/oxfordjournals.molbev.a040454.
Šantl-Temkiv T, Amato P and Casamayor EO et al., (2022). Microbial ecology of the atmosphere. FEMS Microbiology Reviews. 46(4). fuac009. https://doi.org/10.1093/femsre/fuac009.
Sharma AK, Kikani BA and Singh SP (2020). Biochemical, thermodynamic and structural characteristics of a biotechnologically compatible alkaline protease from a haloalkaliphilic, Nocardiopsis dassonvillei OK-18. International journal of biological macromolecules. 153: 680-696. https://doi.org/10.1016/j.ijbiomac.2020.03.006.
Sharma AK, Kikani BA and Singh SP (2021). Diversity and Phylogeny of Actinomycetes of Arabian Sea Along the Gujarat Coast. Geomicrobiology Journal. 38(4). 347-364. https://doi.org/10.1080/01490451.2020.1860165.
Sikorski J, Baumgartner V and Birkhofer K et al., (2022). The evolution of ecological diversity in Acidobacteria. Frontiers in Microbiology. 13(715637). 78. https://doi.org/10.3389/fmicb.2022.715637.
Sorokin DY, Elcheninov AG and Khijniak TV et al., (2022). Natronosporangium hydrolyticum gen. nov., sp. nov., a haloalkaliphilic polyhydrolytic actinobacterium from a soda solonchak soil in Central Asia. Systematic and Applied Microbiology. 45(3). 126307. https://doi.org/10.1016/j.syapm.2022.126307.
Sysoev M, Grötzinger SW, Renn D et al., (2021). Bioprospecting of novel extremozymes from prokaryotes the advent of culture-independent methods. Frontiers in Microbiology. 12. 630013. https://doi.org/10.3389/fmicb.2021.630013.
Thumar JT and Singh SP (2009). Organic solvent tolerance of an alkaline protease from salt-tolerant alkaliphilic Streptomyces clavuligerus strain Mit-1. Journal of Industrial Microbiology and Biotechnology. 36(2). 211. https://doi.org/10.1007/s10295-008-0487-6.
Upadhyay KH, Vaishnav AM and Tipre DR et al., (2019). Diversity assessment and EPS production potential of cultivable bacteria from the samples of coastal site of Alang. Journal of Microbiology, Biotechnology and Food Sciences. 6(1). 661-666. https://doi.org/10.15414/jmbfs.2016.6.1.661-666.
Vaishnav D, Suthar J and Oza T et al., (2014). A statistical approach for the enhanced production of thermostable alkaline protease showing detergent compatibility activity from Bacillus circulans. Biocatalysis and Biotransformation. 32(3):151-160. https://doi.org/10.3109/10242422.2014.913579.
Vera-Gargallo B and Ventosa A (2018). Metagenomic insights into the phylogenetic and metabolic diversity of the prokaryotic community dwelling in hypersaline soils from the Odiel saltmarshes (SW Spain). Genes. 9(3): 152-174. https://doi.org/10.3390/genes9030152.
Vijayaraghavan P, Jebamalar TRJ and Vincent SGP (2012). Biosynthesis optimisation and purification of a solvent stable alkaline serine protease from Halobacterium sp. Annals of microbiology. 62(1): 403-410. https://doi.org/10.1007/s13213-011-0276-8.
Yadav S and Patil SA (2020). Microbial electroactive biofilms dominated by Geoalkalibacter spp. from a highly saline–alkaline environment. npj Biofilms and Microbiomes. 6(1). 1-10. https://doi.org/10.1038/s41522-020-00147-7.
Yang M, Xue Q and Zuo Z et al., (2022). Aliidiomarina halalkaliphila sp. nov., a haloalkaliphilic bacterium isolated from a soda lake in Inner Mongolia Autonomous Region, China. International Journal of Systematic and Evolutionary Microbiology. 72(3). 005263. https://doi.org/10.1099/ijsem.0.005263.
Zhang W, Luo Q and Zhu Y et al. (2018). Microbial diversity in two traditional bacterial douchi from Gansu province in northwest China using Illumina sequencing. PLOS ONE. 13(3). (0194876). 1-16. https://doi.org/10.1016/j.fbio.2019.03.006.


