1Department of Zoology, Kakatiya University, Warangal, Telangana, India.
2 Department of Biotechnology, National Institute of Technology Warangal, Telangana, India.
Corresponding author email: vodithalashamitha@gmail.com
Article Publishing History
Received: 05/04/2021
Accepted After Revision: 10/06/2021
Antheraea mylitta Drury silkworm, a polyphagous, Sericigenous, lepidopteran insect commonly known as tasar silkworm is a species, found in form of 44 ecoraces, feeding on a number of food plants. In India, it covers more than twenty states as ecoraces, with variations in phenotypic traits like fecundity, voltinism, cocoon weight, silk ratio etc. The distribution of the species has encountered diverse geographic and climatic variations of the distinct areas, leading to marked differences in not only phenotypical and physiological traits but also in the commercial and technological aspects. A. mylitta Drury (Andhra Local ecorace), which is an exclusive ecorace of the states of Andhra Pradesh and Telangana, is well known for its superior commercial characters, but, is on the verge of extinction due to its weaknesses in voltinism, emergence, hatching, low yield etc. The ecorace conservation is essential to utilize their valuable genes in enhancing productivity and to build variation in new population through hybridization. Modern sequencing methods like Next-Generation Sequencing technologies and in silico analysis are used in population genetic studies to investigate the evolutionary forces affecting genetic variation. In the present studies, the genomic DNA of parental ecoraces – Andhra local and Daba TV of A. mylitta and their hybrid populations were sequenced independently using the Illumina NextSeq500 in order to analyze their genetic relationship. The sequencing library revealed that the fragment size ranged between 200bp to 700bp and identified 35877 sites in 8 samples. Further, the phylogenetic tree showed closely and distantly related taxa among the populations.
Antheraea mylitta, Ecoraces, Next-Generation Sequencing, Phylogenetic Tree, Variations.
Gattu R, Perugu S, Gangupanthula S. Next-Generation Sequencing (NGS) in the Population of Indian Tropical Tasar Silkworm-Antheraea Mylitta. Biosc.Biotech.Res.Comm. 2021;14(2).
Gattu R, Perugu S, Gangupanthula S. Next-Generation Sequencing (NGS) in the Population of Indian Tropical Tasar Silkworm-Antheraea Mylitta. Biosc.Biotech.Res.Comm. 2021;14(2). Available from: <a href=”https://bit.ly/2T0a1lR“>https://bit.ly/2T0a1lR</a>
Copyright © Gattu et al., This is an open access article distributed under the terms of the Creative Commons Attribution License (CC-BY) https://creativecommns.org/licenses/by/4.0/, which permits unrestricted use distribution and reproduction in any medium, provide the original author and source are credited.
INTRODUCTION
Antheraea mylitta, a wild sericigenous insect is a species widely distributed in the form of 43 ecoraces from West Bengal in the East to Karnataka in the South with its natural inhabitation in the forest areas of Andhra Pradesh, Bihar, Orissa, Madhya Pradesh, Maharashtra, and Telangana. It is a polyphagous insect feeding on a number of food plants primarily on Terminalia arjuna and T. tomentosa, and a host of secondary food plants like Ziziphus, Tectona, Bauhinia, Lagerstroemia, etc. A wide range of distribution of the species has encountered diverse geographic and climatic variations of the distinct areas, leading to marked differences in not only phenotypical and physiological traits but also in the commercial and technological aspects.
Among fifty races of sericigenous insects, twenty-five races of A. mylitta widely available (Jolly et al. 1974). A. mylitta Drury (Andhra Local ecorace), is an exclusive ecorace of the state of Andhra Pradesh, Telangana and is well known for its superior commercial characters. However, it is on the verge of extinction due to its weaknesses in voltinism, emergence, hatching, low yield etc. (Thangavel, 1992; Srivastava et al. 2003; Kavane, 2009). The ecorace conservation is essential for utilizing their valuable genes in enhancing productivity and to build variation in new population through hybridization (Kumaresan et al. 2004; Mirhoseini et al. 2004; Kavane, 2009).
Andhra local ecorace of A. mylitta D. possesses superior commercial cocoon traits, but its commercialization could not be undertaken due to weak voltinism, asynchronized moth emergence (35-40%), poor fecundity (165-205), less amenable to human handling and heavy crop loss during late age silkworm rearing stages resulting in low productivity (10-12 cocoons/dfl). However, this genetic resource material is bestowed with superior commercial characters like low denier (7%), high reliability (66%) and higher silk ratio (16.8%) (Vestergaard et al. 2021). Daba is an adapted ecorace of A. mylitta D. with commercial exploitation under the tasar seed sector. This ecorace shows sustenance in cultivating breeding pocket with moderate fecundity (200-250), S.R. % (13.50-14.50%) and good survival (50-60 cocoons/dfl). Hence, the breeding programme has been formulated involving two ecoraces viz., Daba TV and Andhra local ecoraces. This work aims at the improvement of Andhra local ecorace with bestowed with superior characters i.e., survivability and fecundity of Daba TV (Vestergaard et al. 2021).
The breeding of Tasar Silkworm A. mylitta D (Andhra Local and Daba-TV) has a rich genetic resource as 44 ecoraces, however, the Tasar culture, an important co-discipline of applied forest biology, needs special understanding and addressing towards breeding perspective to promote the sustainable utilization of this precious natural resource (Manohar et al. 2010). Earlier reports revealed that the genetically pathetic characters of this ecorace could be overcome if methodical breeding activities are undertaken probing into the binding capacities and combining abilities of A. mylitta. Identification of several potential markers that contribute to develop genetic characteristics of silkworm population and reveal genetic divergence within low and high yielding strains could have potential practical utility in prospective silkworm breeding program (Vestergaard et al. 2021).
This present work undertaken characterizes the ecoraces of Tasar silkworm, A. mylitta, from different parts of tropical forest zones, as a basis for identification and genetic diversity among the tasar populations. Based on these reports, a comprehensive breeding programme could be evolved to conserve the dwindling population of Tasar silkworm, A. mylitta, Andhra local ecorace. The application of next-generation sequencing (NGS) technology has led to remarkable advances in whole genome sequencing, which provides ultra-throughput sequences to revolutionize plant genotyping and breeding (Jiangfeng et al. 2014). NGS analysis are in constant demand, thus alignment tools and variant calling tools are still improving and are an active area of research (Vestergaard et al. 2021). Further, NGS usage has also been extended to large crop plants, like maize and wheat, to sequence multiplexed samples that combine molecular marker discovery and genotyping. GBS is a novel application of NGS protocol for discovering Single Nucleotide Polymorphism in plant crop populations.
MATERIAL AND METHODS
For the collection of Tasar Silkworm A. mylitta D (Andhra Local and Daba-TV) the parental stocks of ecoraces viz., Andhra Local and Daba TV of Tasar Silkworm A. mylitta raised during the seed crop rearing season, July-August. The Andhra local and Daba TV seed cocoons were collected from RTRS (Warangal district) and Telangana State Silk Board (Chennur Mandal, Adilabad district and Mahadevpur, Karimnagar districts) respectively (Fig. 1.a, b). They arranged in the form of garlands in grange chambers. The disinfection of room is using with 2% formaldehyde, prevented by arranging suitable nylon net, ventilated grainage chambers in the laboratory. The date of emergence of each of the ecorace (male/female) noted. The male and female moths emerged out of Non – diapause cocoon stocks of the above divergent geographic ecoraces used for the study.
Figure 1: A. Collection of seed cocoons Telangana State Silk Board; B. Daba- TV Seed cocoons.

The moth emergence was studied during the first grainage season (June-July). The emergence of moths starts late in the evening mostly, between 18.00 to 22.00 hr. Coupling of moths just after emergence is not allowed. By keeping male and female moths separately in the cages for about 2-3 hr so as to from 19.00 hr and continued u to 23.00 hr. The number of moths emerged and male-female synchronization was recorded in Andhra local and Daba-TV parental ecoraces. The male moths start emerging 1- 2 days earlier than female moths. Immediately after emergence male-female moths were selected from different batches as per the requirement, to prepare backcrosses among Andhra local and Daba-TV ecoraces (Fig 2).
Figure 2: Moth emergence of (Andhra local and Daba TV) Tasar Silkworm, A. mylitta D.

For the backcross methodology, the backcross breeding of silkworm using parents with preferred traits and selection in subsequent generations offer superior varieties. The outcrossing or repeated backcrossing with exotic races can improve targeted character(s) and induction of polyvoltine traits into uni/ bivoltine enhances resistance in breeds the retention of 50% of superior economic traits of in-situ grown wild ecorace in F1 generation and subsequent attainment of 75%, 87.5% and so on of domesticated blood in BC1, BC2 and so on progenies makes their stock maintenance easy, and to backcross with amenable Daba ecorace than choosing a wild ecorace (in-situ or ex-situ) as donor (recurring parent) for backcrossing (Aruga, 2001).
Breeding of silkworm means the improvement of silkworm strains, which is an extremely important issue for the silk industry. Methods adopted for the improvement of silkworm strains are more or less the same as those in other animals and plants. Three methods are followed viz., (1) selection of pure lines, (2) cross-breeding, and (3) induction of mutations. At present, out of the three methods, cross-breeding is the most popular and widely applied to develop a large number of strains. In cross-breeding, in most cases, an objective is set and decided upon and strains suitable for the purpose are carefully selected. By producing hybrids of the two parent strains, various combinations of characters manifested in the filial generations are observed and the excellent ones are selected and fixed (Aruga, 2001). In the present study, the breeding of tasar ecoraces was done as follows: The preparation of F1 hybrids was done by crossing Daba TV (donor/female) and Andhra local (recipient/ male) which is aimed at the introgression of survivability traits.
Crosses were made by releasing male and female moths in the ratio of 2:4 under in situ conservation model The dfls of F1 was prepared and incubated in the laboratory.The F1 progeny was brushed and reared under ex-situ conditions during the first crop season.Parental stocks of Daba and Andhra local were maintained in separate locations to prevent chances of intermixing of genetic characters. The F1 hybrid male (Daba TV X Andhra local) was backcrossed with the female of Andhra local to produce the back-cross dfls in F2.Incubation of back-cross dfls was carried out in the laboratory.The F2 progeny was brushed and reared under ex-situ conditions during the second crop season.Observations and interpretations were noted down.
Production of F1 and F2 Hybrids: During hybridization, Daba TV (female) was crossed with Andhra local (male) to produce F1 hybrids, in which Daba TV (female) will be donor parent whereas male counterpart of Andhra local acts as recipient parent. F1 (male) was crossed with Andhra local (female) to produce F2 hybrids. Rearing performance of different backcrosses was observed for different parameters and recorded as follows.
During hybridization, in the next crop, i.e., during September, F1 (male) was crossed with Andhra local (female) to produce F2 hybrids, in which F1 (male) will be donor parent whereas male counterpart of Andhra local acts as recipient parent. This method is aimed at introgression of survivability traits of Andhra local ecorace. Genomic DNA isolation: The Genomic DNA isolation and the quantification of the 1-8 samples of Tasar Silkworm, A. mylitta D Parental ecoraces (Andhra local, Daba TV, F1 and F2) of each line by using HiPurATM Insect DNA Purification Kit.
DNA was dissolved in TE buffer (Tris-EDTA, pH 8.0), revealed an ideal concentration of DNA ng / µl of the genomic sample i.e., between 1.8 – 1.9 in 260 / 280 ratio) checked against 1 kb standard DNA ladder was obtained. It been observed that DNA has been isolated without any protein or any other contamination and was used for further studies in PCR and NGS analysis. Genetic characterization of 4 populations viz., parental ecoraces (Andhra local, Daba TV), F1 and F2 were done (Suzuki et al. 1972; Nagaraja and Nagaraju 1995; Aruga, 2001).
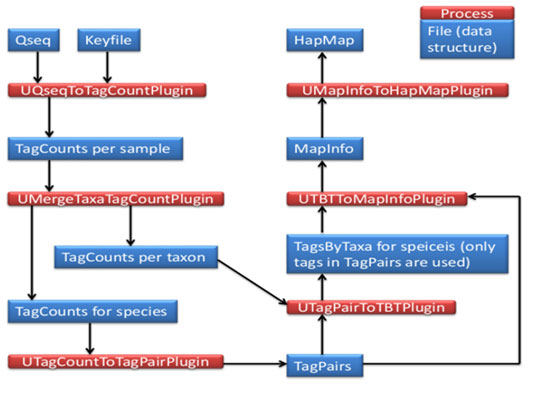
Database & Tools used for analysis of sequence: The genomic DNA of A. mylitta was sequenced independently using the Illumina NextSeq500; 75 paired-end and data analysis were done by FastQC1, bowtie2, and Tassel-33 respectively. Total no of sample sequences is 8 and PstI-HF – MluC1 enzymes were used. Later, the sequence was Converted using tagCount, this plugin derives a tagCount list for each sample. It keeps only good reads having a barcode and a cut site. Trims off the barcodes and truncates sequences that have a second cut site, or read into the common adapter. In the current studies, Tassel3 UNEAK pipeline, a denovo GBS analysis approach was opted due to the lack of reference genome. The entire protocol (Fig 3 and 4) is explained step by step in the following sections.
Figure 3: Experimental Work flow for Genotyping by Sequencing Library Preparation.
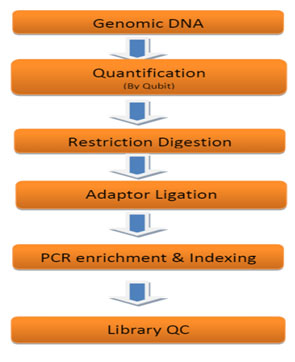
Figure 4: Schematic Overview of denovo GBS analysis workflow.
Library Preparation and Sequencing: GBS Library Preparation was performed at Genotypic Technology’s Genomics facility. 300 ng of Qubit quantified genomic DNA was used for double digestion by Pst-HF and MluCl restriction enzyme at 37°C for 4 hours. The resulting digested fragments were directly ligated to pair of restriction site-specific adapters at 20°C for 1hour using T4 DNA ligase. The adapter ligated fragments were subjected to PCR amplification (10 cycles) to amplify the adapter-ligated fragments and to add sample specific dual index barcodes (Nextera XT v2 index kit, Illumina, U.S.A.). The amplified Illumina-compatible sequencing library was quantified by Qubit fluorimeter (Thermo Fisher Scientific, MA, USA) and its fragment size distribution was analyzed on Agilent 2200 TapeStation (Table1).
Table 1. Genomic DNA samples of parental and hybrid ecoraces of Tasar Silkworm A. mylitta D and their indexes.
| # | Sample ID | Index1 | Index1 Sequence | Index2 | Index2 Sequence |
| 1 | SO_7874_Repl_S1 | N701 | TAAGGCGA | S507 | AAGGAGTA |
| 2 | SO_7874_Repl_S5 | N702 | TAAGGCGA | S507 | AAGGAGTA |
| 3 | SO_7874_Repl_S7 | N703 | TAAGGCGA | S507 | AAGGAGTA |
| 4 | SO_7874_Repl_S8 | N704 | TAAGGCGA | S507 | AAGGAGTA |
| 5 | SO_7874_Repl_S6 | N705 | TAAGGCGA | S508 | CTAAGCCT |
| 6 | SO_7874_S2 | N706 | TAAGGCGA | S508 | CTAAGCCT |
| 7 | SO_7874_S3 | N707 | TAAGGCGA | S508 | CTAAGCCT |
| 8 | SO_7874_S4 | N710 | TAAGGCGA | S508 | CTAAGCCT |
The materials which are used; TaKaRa Ex TaqTM (TaKaRa), Pstl-HF restriction enzyme (NEB), MluC1 restriction enzyme {(NEB), T4 DNA Ligase}, HighPrep PCR (Magbio), Nuclease Free Water (Ambion), D1000 Tape station kit (Agilent), Qubit DNA HS kit (Invitrogen), Nextera XT index kit v2-set D (Illumina).
RESULTS AND DISCUSSION
The sequencing library for all the samples are suitable for Illumina sequencing studies and found that the fragment size ranged between 200bp to 700bp. As the combined adapter size is approx. 120bp, the effective user- defined insert size is 80bp to 580bp (Table1). The GBS analysis of 8 samples revealed the related information of taxa; gametes, Number of Gametes, Gametes not missing, Proportion Gametes not missing, Gametes missing; heterozygotes- number and proportion. The results are shown in (Table 2).
Table 2. Complete summary of the Taxa of parental and hybrid ecoraces of Tasar Silkworm A. mylitta D.
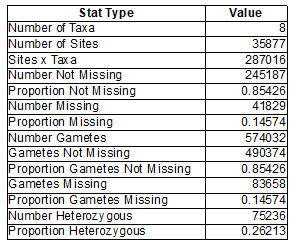
The GBS analysis of 8 samples revealed the related information of taxa; gametes, Number of Gametes, Gametes not missing, Proportion Gametes not missing, Gametes missing; heterozygotes- number and proportion. The results are shown in (Table 2). The GBS analysis of 8 samples revealed the related information of taxa; gametes, Number of Gametes, Gametes not missing, Proportion Gametes not missing, Gametes missing; heterozygotes- number and proportion. The results are shown in (Table 2). GBS sequencing analysis showing no. of alleles with their proportions are identified with respective nucleotides and amino acids (Table 3).
Table 3. Summary of Alleles of parental and hybrid ecoraces of Tasar Silkworm A. mylitta D.
| Alleles | Number | Proportion | |
| C | 45181 | 0.15742 | 0.18427 |
| G | 43408 | 0.15124 | 0.17704 |
| N | 41829 | 0.14574 | 0.1706 |
| T | 41490 | 0.14456 | 0.16922 |
| A | 39872 | 0.13892 | 0.16262 |
| Y | 24637 | 0.08584 | 0.10048 |
| R | 23574 | 0.08213 | 0.09615 |
| W | 9054 | 0.03155 | 0.03693 |
| M | 6716 | 0.0234 | 0.02739 |
| K | 6112 | 0.02129 | 0.02493 |
| S | 5143 | 0.01792 | 0.02098 |
The sequenced 8 samples and identified their populations with related taxons. Moreover, the identified SNPs at the same location in all the sequences were found. When cluster analysis was performed, all the eight samples were same in length and clustered together at identical in sequence length i.e., the range of 4 – 9. The eight sequences have shown unique GC rich regions (Table 4).
Table 4. Information for the eight benchmark sequences and sequences Statistical analysis data of parental and hybrid ecoraces of Tasar Silkworm A. mylitta
| sample
method |
S1 | S2 | S3 | S4 | S5 | S6 | S7 | S8 |
| Sequence length | 75 | 75 | 75 | 75
|
75 | 75 | 75 | 75 |
| GC% | 36 | 35 | 36 | 36 | 36
|
35 | 35 | 33 |
| Phrad Score | 34 | 34 | 34 | 34 | 34 | 34 | 34
|
34 |
| Clustered | 6-8
|
4-9 | 4-9 | 3-9
|
4-9 | 4-9 | 5-9 | 4-8 |
| Kmer | 1-5 | 1-5
|
1-5 | 1-5 | 1-5 | 1-5 | 1-5 | 1-5 |
| Total Sequence | 2644277 | 2877078 | 3365729 | 2380003
|
3330948 | 3948642
|
2984491 | 1376395
|
GC content (guanine-cytosine content) was found to be high in S1, S5 and S6, compared to S2, S3, S4 and S7, However, a species with an extremely low GC–content was S8 (Fig.5 and 6). The figure 7 shows the comparative analysis of 8 sequences. From the sequence analysis it was found that some of the amino acid regions were aligned together as shown in green colour.
Figure 5: Distribution of GC percentage.
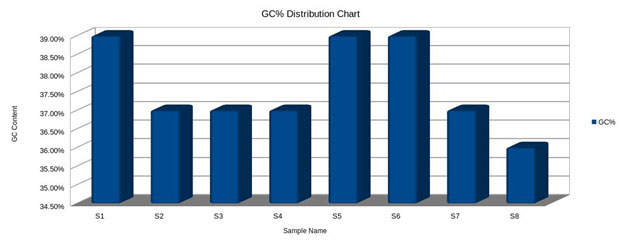
Figure 6: Sequences Read distribution.
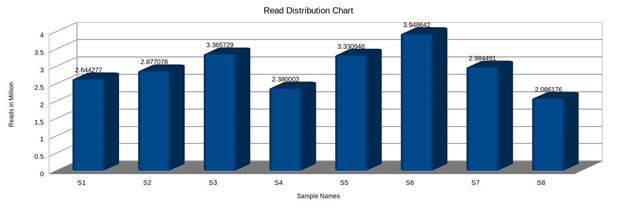
Figure 7:Eight samples DNA sequence as highlighted basis on their amino acid function.
The Phylogenetic tree has shown formation of Three clusters. F1 and F2 parents have formed individual clusters. Parental ecoraces and populations formed clusters (Fig 8).
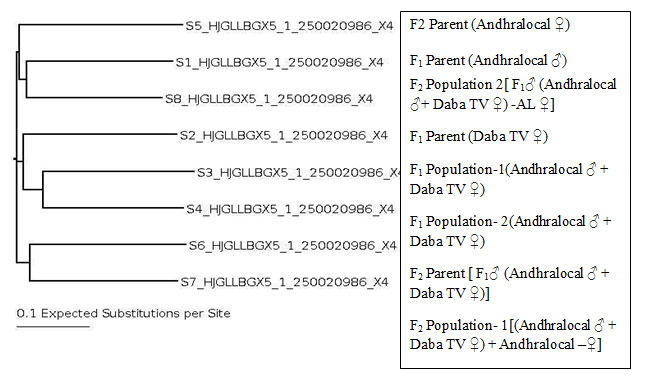
F1 parent & F2 population formed one cluster, F1 parent & F1population formed one cluster, F2 parent & F2 population formed one cluster. The molecular characterization and phylogenetic analysis revealed parental ecoraces and populations formed one cluster, while F2 Parent and F1 Parent formed individual clusters. It may be noted that the F2 population is genetically closer to the parental cluster (AL male-DTV female) than that of F1. It is also interesting to note that F2 population is closer to F1 parent, that is Andhra local male.
NGS protocols for discovering and genotyping SNPs for crop improvement. The low cost of GBS makes it an attractive approach to saturate the mapping and breeding populations with a high density of SNP markers. Successive improvements of the sequencing chemistries and base-calling software will allow NGS technologies to deliver higher sequencing throughputs per run, which in turn enables deeper multiplexing for a fixed average sequencing depth per sample (Elshire et al. 2011; Poland et al. 2012).
As the sequence-based genotyping is available for a whole range of genomic studies, GBS will stand to be one of the major components in silkworm genetics and breeding. Even though the availability of reference genomes is very useful to eliminate repetitive sequences, GBS can be used without a reference genome, by either using consensus sequences of reads as the reference or using the tags simply as dominant markers (Elshire et al. 2011). In addition, GBS markers can be done with extremely high accuracy, with a reference genome in biparental mapping populations, allowing for low coverage of the offspring at an even lower cost, after genotyping parents and their offsprings at high coverage (Elshire et al. 2011; Poland et al. 2012).
The sequencing of 8 samples in the present investigation has identified their populations with related to taxons and identified the SNPs at the same location in all the sequences. The cluster analysis of the eight samples has shown that they were of same length and clustered together at i.e., the range is 4 – 9. All such sequences have also shown unique GC rich regions (Poland et al. 2012).
GC content (guanine-cytosine content) is found to be high in S1, S5, and S6 compared to S2, S3, S4, and S7. However, a species with an extremely low GC–content is S8. Most NGS platforms are able to generate reliable sequences and display near perfect coverage behavior on GC-rich, neutral and moderately AT-rich genomes. However, there are key differences between the quality of that data and the applications it will support. Illumina is less predisposed to homopolymer errors, it shows an overall accuracy greater than 99.5%, but sometimes can provide under-representation of regions (i.e., AT/GC-rich) and nucleotide substitution errors (Bentley et al. 2008; Dohm et al. 2008; Harismendy et al. 2009; Nakamura et al. 2011; Quail et al. 2012).
We applied the nine trimming algorithms on four different datasets (see Materials and Methods). The quality of these datasets was assessed with FastQC (see File S1 and Figure S1 for Q distribution plots) and measured by different metrics, such as the average PHRED error score, GC content biases and position-specific quality variations. The datasets vary conspicuously, possessing almost perfect quality parameters for the Yeast DNA-Seq dataset and somehow average-to-high for Lovell raw reads (Figure S1).
CONCLUSION
The sequencing of 8 samples in the present investigation has identified their populations related to taxons and identified the SNPs at the same location in all the sequences. The cluster analysis of the eight samples has shown that they were of same length clustered together at i.e., the range is 4 – 9. All such sequences have also shown unique GC rich regions. GC content (guanine-cytosine content) is found to be high in S1 (Andhralocal (male) –F1 Parent), S5(Andhralocal (Female) – F2 Parent), and S6(F1 (male) – – F2 Parent) compared to S2(Daba TV (Female) – (F1 Parent), S3 (F1 Population1), S4(F1 Population 2) and S7(F2 Population 1). However, species with an extremely low GC-content is S8. The conservation of Tasar silkworm, A. mylitta D, Andhra’s local ecorace needs further improvement on breeding practices, to enable the viability of F1 and F2 generations. The present investigation on PCR based Phylogenetic analysis using Mega Software in 4 tasar populations viz., parental ecoraces [(Andhra local, Daba TV), F1 and F2], derived from hybridization between two contrasting genetically variable ecoraces viz., of Andhra local and Daba TV was successful for two successive commercial crops from commercial viewpoint.
ACKNOWLEDGEMENTS
Authors are thankful to University Grants Commission (New Delhi), for providing funds required to carry out the present work under Ref: Lr.No.F.15-1/2016-17/PDFWM-2015-17-TEL-31108(SA-II), dt.11-04-2016.
Conflict of Interests: The authors declare no competing interests among themselves while conducting the research and preparing the report.
REFERENCES
Bentley DR, Balasubramanian S, and Swerdlow H.P (2008) Accurate whole human genome sequencing using reversible terminator chemistry. Nature, 456(7218):53-9. https://doi. org/10.1038/nature07517
Dohm JC, Lottaz C, and Borodina T (2008) Substantial biases in ultra-short read data sets from high-throughput DNA sequencing. Nucleic Acids Res, 36(16):e105. https://doi.org/10.1093/nar/gkn425
Elshire RJ, Glaubitz JC, and Sun Q (2011) A robust, simple genotyping-bysequencing (GBS) approach for high diversity species. PLoS ONE, 6:e19379. doi:10.1371/journal.pone.0019379
Harismendy O, Ng PC, and Strausberg RL (2009) Evaluation of next generation sequencing platforms for population targeted sequencing studies. Genome Biol, 10(3):R32. https:// doi.org/10.1186/gb-2009-10-3-r32
Aruga H (2001) Principles of Sericulture, Oxford and Ibh Publishing co. Pvt. Ltd.
He J, Zhao X, and Laroche A (2014) Genotyping –by-sequencing (GBS), an ultimate marker- assisted selection (MAS) tool to accelerate breeding Frontiers in plant Science Plant. Genetics and Genomics, 4(484):1-8.
Jolly MS, Sen SK, and Ashan MM (1974) Tasar culture. Ambika publishers Bombay, 134-147.
Kavane RP, and Sathe TV (2009) On a new variety (kolhapurensis) of Antheraea mylitta from India Biological forum. An international Journal, 1(2):5-47.
Kumaresan P, Sinha RK, and Mohan B (2004) Conservation of mutivoltine silkworm (Bombyx mori L.) Germplasm in India – an overview. International Journal of Industrial Entomology, 9(1):1-13.
Vestergaard LK, Oliveira DNP, and Høgdall CK (2021) Next Generation Sequencing Technology in the Clinic and Its Challenges. Cancers, 13:1-18. https://doi.org/10.3390/cancers13081751
Reddy MR (2010) Breeding perspective for silk yield and Quality in Indian Tropical Tasar Silkworm, Antheraea mylitta D (Lepidoptera: Saturniidae). Journal of Applied Science, 10(7):1902-1909.
Mirhoseini SZ, Seydavi AR, and Ghanipoor M (2004) General and special combining ability estimation and heterosis in new varieties of silkworm Bombyx mori. Journal of Biological Science, 4:725-730.
Nagaraju GM, and Nagaraju J (1995) Genome fingerprinting of the silkworm Bombyx mori using random arbitrary primers. Electrophoresis, 16:1633-1638.
Nakamura K, Oshima T, and Morimoto T (2011) Sequence-specifc error profle of Illumina sequencers. Nucleic Acids Res, 39(13):e90. https://doi.org/10.1093/nar/gkr344
Poland JA, Brown PJ, and Sorrells ME (2012) Development of high-density genetic maps for barley and wheat using a novel two-enzyme genotyping-by-sequencing approach. PLOS ONE, 7(2): e32253-e19379.
Quail MA, Smith M, and Coupland P (2012) A tale of three next generation sequencing platforms: comparison of Ion Torrent, Pacific Biosciences and Illumina MiSeq sequencers. BMC Genomics, 13341:10.1186/1471-2164-13-341.
Srivastava AK, Sinha AK, and Sinha BRRP (2003) Descriptor of topical tasar silkworm, Antheraea mylitta Drury (Lepidoptera: Saturniidae), CTR&TI Ranchi, India.
Suzuki Y, Gage L, and Brown DD (1972) The genes for silk fibroin in Bombyx mori. J. Mol. Biol, 70:637–649.
Thangavelu K (1991) Recent studies in Indian tasar and other wild silkmoths. Wild Silkmoth, 20-29.
Thangavelu K (1992) Population ecology of A. mylitta Drury (Lepidoptera Saturnidae). Wild Silkmoths, 92:99-104.


