1Department of Biology, College of Science, University of Jeddah, Jeddah 21589, Saudi Arabia
2Departement of Food Hygiene, Faculty of Veterinary Medicine, New Valley University, Egypt.
orresponding author email: dr.nagwa2004@yahoo.com
Article Publishing History
Received: 19/07/2021
Accepted After Revision: 25/09/2021
Red meat consumption (chevon, beef, and lamb) is one of the most traditional favorable dishes and remain in the modern lifestyle. Chevon is the meet of adult goats and it is considered one of the beef and sheep meat competitor. It is characterized by its lower calories content in comparison to beef content, total fat; “cholesterol and saturated fat” which maintain consumers health. Studies on chevon-microbial interactions are not fulfilled globally, therefore, we aimed to achieve microbial survey and molecular identification of chevon meat food contaminants from different markets of Makkah region, Saudi Arabia. A total of 50 chevon samples were purchased from different retail markets within Makah from September 2019 until January 2020. Samples were transported to the laboratory in a cooler.
They were macerated in peptone water and then cultured on selective media of some indicator’s microorganisms. About; 2.1×105, 2.1×104, 2.1×105, 1.2×10, 1.5×105, 2.5×105, 1.6×105, 3.7×105 CFU/g were the mean of total aerobic counts, anaerobic count, Enterobacteriacea spp. and Staphylococcus spp. counts respectively. All tested chevon meat samples were within permissible limit and fit for human consumption. Molecular Identification of isolated microorganism were as following; Staphylococcus sciuri were 2/9 (22.2), while; for all, Aeromonas enteropelogenes, Escherichia fergusonii, Proteus mirabilis, Pseudomonas aeruginosa and Staphylococcus lentus were 1/9 (11.1) respectively. This is the first report isolated of Aeromonas enteropelogenes from chevon meat. All isolated microorganisms had public health concern as food poisoning microorganisms. Further investigation needed to study the chevon meat and its microbial quality.
Aeromonas Enteropelogenes, Foodborne Microorganisms, Goat
Meat, 16s Rdna, Proteus Mirabilis, Pseudomonas Aeruginosa.
Allhyani A.A, Baeshen M, Elsharawy N.T. Molecular Identification of Newly Isolated Foodborne Bacterial Strains from Chevon Meat. Biosc.Biotech.Res.Comm. 2021;14(3).
Allhyani A.A, Baeshen M, Elsharawy N.T. Molecular Identification of Newly Isolated Foodborne Bacterial Strains from Chevon Meat. Biosc.Biotech.Res.Comm. 2021;14(3). Available from: <a href=”https://bit.ly/3fT9gDO“>https://bit.ly/3fT9gDO</a>
Copyright © This is an Open Access Article distributed under the Terms of the Creative Commons Attribution License (CC-BY). https://creativecommons.org/licenses/by/4.0/, which permits unrestricted use distribution and reproduction in any medium, provided the original author and sources are credited.
INTRODUCTION
Red meat consumption (chevon, beef, and lamb) is one of the most traditional favorable dishes and remain in the modern lifestyle (Jiang and Xiong, 2016). The goat meat (Chevon) is very common and widely flavored by the consumers worldwide. Chevon is a type of red meat but better in their nutritional vales when compared with beef and other red meat types. Unlike with beef chevon is lower in calories, cholesterol and saturated fat which protect the consumers health especially for consumers which suffering from heart disease that affected by the lower sodium and higher potassium content in addition to their essential amino acids which differ than other red meats, the chevon cutlets consider as a better option for consumers (Singh et al., 2014). Recently, chevon consumption become more attractive to the consumers due to the risen of the health conscious as this type of meat characterized by its lower fat contents than other different red meat types which make it the most excellent low fat meat sources (Mazhangara et al., 2019 and Sial, et al., 2021).
Three mechanisms in different meat products spoilage occur during processing and/or storage: lipid oxidation, enzymatic autolysis, and microbial spoilage. Microbial population affected by microflora of the animals, skin and intestinal tract, in addition to other sources of microbial contamination handling, from environment, storage conditions. Growth of microorganisms in meat appeared as slime formation, off odors, texture, changes in appearance, degradation of components and change the water holding capacity. Oxidation of lipid affected by the composition of different fatty acids, vitamin E content, and free iron content in muscles.
Many enzymes degrade different nutrient component such as proteins, fats and carbohydrates of the tissues which resulting in greenish discoloration and softening of meat which resulting in microbial decomposition. The proteolytic enzymes remain active even at low temperatures (5∘C) which leading to loss of water holding capacity, microbial growth, and biogenic amines production (Heifa’a et al., 2018 and Vita, et al., 2020; Priya et al., 2021).
Food borne pathogens considers the main problems, especially in developing countries. Food considered as the most important sources that causes the microorganisms to human, these microorganisms still a major cause of food-borne human disease in most parts of the world (Aseel et al., 2011). The presence of food pathogens in food items mainly leading to undesirable gastrointestinal symptoms. It has been mentioned that hazards such as bacteria, fungi, allergens, chemicals, and foreign matter can be present in different meat types of bacterial pathogens (Pal et al., 2018 and Abebe, et al., 2020). There was a big shortage in chevon microbial evaluation studies globally that is the reason of our choice and aim of this study which conducted to survey the microbial contamination and serological identification of chevon meat from different markets in Makkah region, Saudi Arabia.
MATERIAL AND METHODS
A total of 50 chevon samples, about 450g/each were purchased from different retail markets within Makah from September 2019 until January 2020. Samples were collected and shipped without delay in pre-cooled insulated containers with frozen packs to laboratory. All samples were prepared according to the technique recommended by APHA (2002) as following, one gram from each sample were homogenized under aseptic condition with 9 ml of sterile buffered peptone water 0.1%, and then homogenized to have a dilution (1/10) further serial dilution up to 107 were prepared. About 0.1 ml of each prepared serial dilution were inoculated on standard plate count agar then incubated at 30±1oC/72±3 hrs. the obtained colonies were recorded.
The same procedures were incorporated for total anaerobic counts using Reinforced Clostridial medium agar (Oxoid; CM151), then were incubated in an anaerobic jar (Gaspak plus anaerobic system) at 37oC/48 hours. Countable plates were recorded. Staphylococcus sp. Count (ISO, 1999) using mannitol salt agar which incubated at 30±1oC/ 24 – 48 ±4 hrs. Suspected yellow colonies with yellow zone were counted and then 5 typical colonies were picked up on nutrient agar slant for further confirmation biochemically by Coagulase test using rabbit plasma, suspected colonies were transferred to Brain heart infusion (BHI) broth tube at 35 – 37oC/24 hrs then 0.1 ml of the culture was aseptically mixed to the sterile test tube containing 0.3 ml rabbit plasma (Difco, BD) at 35–37oC/4hrs, if test was – ve was re-examined each 2 hrs until 24 hrs.
Total Enterobacteriaceae Count (ISO, 2004) by violet red bile glucose agar (VRBGA) at 30-35oC/24 hrs. DNA extraction: suspend a single microbial cell in 20 μl of lysis buffer containing 0.25% (vol/vol) sodium dodecyl sulfate and 0.05 N NaOH. Then heating at 95°C/15 min., addition of chromatography-grade H2O (Fisher), and stored the lysis suspension at −20°C (Spilker et al., 2004). The extraction of DNA done according to manufactured of commercial DNA extraction kit (Presto Mini-DNA Bacteria Kit. Geneaid Biotech Ltd. USA) instructions. Which followed by DNA extracted using nanodrop device at wave length of 260/280 nm (Al-Azawi et al., 2018).
Primer design: 16S rDNA relevant sequences which were presented in the database of the GenBank. Based on this alignment, species-specific primers, and putative genus- primers were designed and shown on table (1) (Spilker et al., 2004). PCR master mix preparation from AccuPower®PCR-PreMix-Kit according to the company directions as shown in (Table 2), then all the PCR tubes were vortexed for 3 min and were transferred in the thermocycler apparatus (MyGene, Bioneer. Korea) (Al-Azawi et al., 2018).
Amplification of targeted DNA was added in 25-μl volume, that containing; 50 mM Trizma (St. Louis, Mo., pH 8.3; Sigma), 2 mM MgCl2, 0.4 μM for each primer, 1U of Taq polymerase (Invitrogen, Carlsbad, Calif.), 250 μM for each deoxynucleoside triphosphates (Promega, Madison, Wis.) and 2 μl whole-cell bacterial lysate, added to 25 μl of high-performance liquid chromatography-grade H2O. Amplification by a RapidCycler (Idaho Technology Inc., Salt Lake City, Utah) thermo controller (Spilker et al., 2004). The determination 16S rDNA sequence: to ensure identification PCR-based results, analyzing 16S rDNA comparative sequence performed.
The complete 16S rRNA genes (positions between 9-1500 by numbering system) which PCR amplified using Pfu DNA polymerase (Stratagene, La Jolla, Calif.) with conserved primers UFPL and URPL as described. DNA purification using QIAquick PCR purification kit (Qiagen Inc., Valencia, Calif.). Sequencing DNA performed using Applied Biosystems ABI model 3700 sequencer (PE Applied Biosystems, Foster City, Calif.) with BigDye Terminator cycle sequencing ready reaction kit. Resultant sequences visualized as chromatograms then manually edited using Chromas version 2.22 (Technelysium Pty. Ltd., Helensvale, Australia). Sequences assembled by EditSeq (DNASTAR Inc.) and identified by BLASTN compared with the NCBI database available sequences (www.ncbi.nlm.nih.gov/BLAST ) (Spilker et al., 2004).
RESULTS AND DISCUSSION
Bacteriological Profile of Chevon Samples revealed on table (2) as following; the positive aerobic bacteria were about 50/50 (100%) of the total chevon sample same result detected in case of Anaerobic, Enterobacteriacea spp. count while, only about 25/50 (50%) of tested chevon sample have Staphylococcus spp.
Table 1. Molecular identification
| Description | Sequencing primer name primer sequences | PCR primer name primer sequences |
| Aeromonas enteropelogenes | 785F 5′ (GGA TTA GAT ACC CTG GTA) 3′ | 27F 5′ (AGA GTT TGA TCM TGG CTC AG) 3 |
| 907R 5′ (CCG TCA ATT CMT TTR AGT TT) 3′ | 1492R 5′ (TAC GGY TAC CTT GTT ACG ACT T) 3′ | |
| Escherichia fergusonii | 785F 5′ (GGA TTA GAT ACC CTG GTA) 3′ | 27F 5′ (AGA GTT TGA TCM TGG CTC AG) 3 |
| 907R 5′ (CCG TCA ATT CMT TTR AGT TT) 3′ | 1492R 5′ (TAC GGY TAC CTT GTT ACG ACT T) 3′ | |
| Proteus mirabilis | 785F 5′ (GGA TTA GAT ACC CTG GTA) 3′ | 27F 5′ (AGA GTT TGA TCM TGG CTC AG) 3′ |
| 907R 5′ (CCG TCA ATT CMT TTR AGT TT) 3′ | 1492R 5′ (TAC GGY TAC CTT GTT ACG ACT T) 3′ | |
| Pseudomonas aeruginosa | 785F 5′ (GGA TTA GAT ACC CTG GTA) 3′ | 27F 5′ (AGA GTT TGA TCM TGG CTC AG) 3 |
| 907R 5′ (CCG TCA ATT CMT TTR AGT TT) 3′ | 1492R 5′ (TAC GGY TAC CTT GTT ACG ACT T) 3′ | |
| Staphylococcus Lentus | 785F 5′ (GGA TTA GAT ACC CTG GTA) 3′ | 27F 5′ (AGA GTT TGA TCM TGG CTC AG) 3 |
| 907R 5′ (CCG TCA ATT CMT TTR AGT TT) 3′ | 1492R 5′ (TAC GGY TAC CTT GTT ACG ACT T) 3′ | |
| Staphylococcus Sciuri | 785F 5′ (GGA TTA GAT ACC CTG GTA) 3′ | 27F 5′ (AGA GTT TGA TCM TGG CTC AG) 3 |
| 907R 5′ (CCG TCA ATT CMT TTR AGT TT) 3′ | 1492R 5′ (TAC GGY TAC CTT GTT ACG ACT T) 3′ |
Table 2. Bacteriological profile of tested chevon samples
| (Bacteriological profile)
|
Positive samples | Negative samples | ||
| No. | Percent % | No. | Percent % | |
| Aerobic bacteria | 50 | 100 | 0 | 00 |
| Anaerobic bacteria | 50 | 100 | 0 | 00 |
| Enterobacteriacae spp. counts | 50 | 100 | 0 | 00 |
| Staphylococcus spp. | 25 | 50 | 25 | 50 |
Statistical analysis for Different Types of Micro-organisms (CFU/g) in the chevon samples discussed on table (3) were the total aerobic counts reported about; 90×103, 38×104, 2.1×105 ±2.1×104 as minimum, maximum, mean ±SE values respectively while, about; 16.2×104, 32.1×104, 2.1×105 ±1.2×10 CFU/g. were detected as minimum, maximum, mean ±SE values respectively in case of total anaerobic count. Enterobacteriacea spp. showed about; 20×103, 6.0×105, 1.5×105 ±2.5×105 CFU/g. as minimum, maximum, mean ±SE values respectively while, about; 10×103, 1.2×106, 1.6×105 ±3.7×105 CFU/g. were detected as minimum, maximum, mean ±SE values respectively in case of Staphylococcus spp. counts.
Table 3. Statistical analysis for Different Types of Micro-organisms (CFU/g) in Chevon samples
| Micro-organisms | Minimum | Maximum | Mean | SE± |
| Total Aerobic counts | 90 X 103 | 38 X 104 | 2.1X 105 | 2.1 X 104 |
| Total Anaerobic count | 16 X 104 | 32 X 104 | 2.1X 105 | 1.2 X 10 |
| Enterobacteriacae spp. | 20 X 103 | 6.0 X 105 | 1.5 X 105 | 2.5 X 105 |
| Total Staphylococcus spp. counts | 10 X 103 | 1.2 X 106 | 1.6 X 105 | 3.7 X 105 |
Comparison with microbiological criteria for foodstuffs (GSO 1016/ 2015): According to Saudi Arabia microbiological criteria for foodstuffs (GSO 1016/2015) the permissible limit of different meat products average between; 5×105-5×106 CFU/gm. In case of total aerobic count and anaerobic, 102-103 CFU/gm in case Enterobacteriacae, all samples should be free from foodborne pathogens, while, in case of Staphylococcus the acceptable limit ranged between 102 -103 CFU/gm. Comparison with microbiological criteria for foodstuffs (GSO 1016/ 2015) viewed in table (4) which declared that; all tested chevon meat samples were within permissible limit and fit for human consumption.
Table 4. Comparison with microbiological criteria for foodstuffs (GSO 1016/ 2015)
| (Bacteriological profile)
|
Within permissible | Over permissible | ||
| No. | Percent % | No. | Percent % | |
| Aerobic bacteria | 50 | 100 | 0 | Zero % |
| Anaerobic bacteria | 50 | 100 | 0 | Zero % |
| Enterobacteriacae spp. counts | 50 | 100 | 0 | Zero % |
| Staphylococcus spp. | 50 | 100 | 0 | Zero % |
Molecular Identification of isolated microorganism: PCR identification mentioned in table (5) and figures (1-6) were as following; Staphylococcus sciuri were 2/9 (22.2), while; for all of, Aeromonas enteropelogenes, Escherichia fergusonii, Proteus mirabilis, Pseudomonas aeruginosa and Staphylococcus lentus were 1/9 (11.1) respectively.
Table 5. Different Incidence of Molecular Identification of Chevon samples
| Bacteriological profile | Positive samples | Negative samples | ||
| No | Percent % | No | Percent % | |
| Aeromonas enteropelogenes | 1 | 11.1 | 8 | 88.8 |
| Escherichia fergusonii | 1 | 11.1 | 8 | 88.8 |
| Proteus mirabilis | 1 | 11.1 | 8 | 88.8 |
| Pseudomonas aeruginosa | 1 | 11.1 | 8 | 88.8 |
| Staphylococcus lentus | 1 | 11.1 | 8 | 88.8 |
| Staphylococcus sciuri | 2 | 22.2 | 7 | 77.7 |
Figure 1: phylogenetic molecular tree of Aeromonas spp. isolate and most relate genera
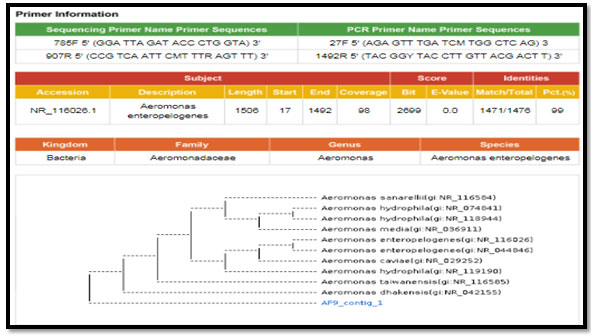
figure 2: phylogenetic molecular tree of Escherichia spp. isolate and most relate genera
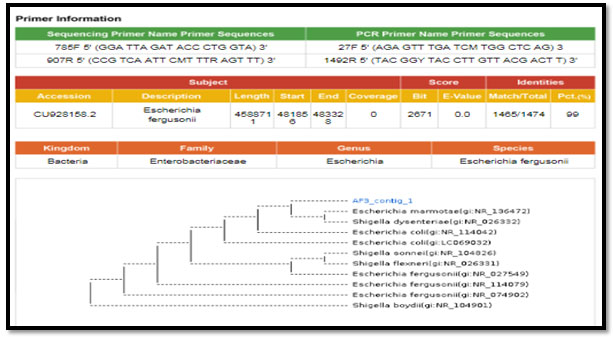
Figure 3: phylogenetic molecular tree of Proteus spp. isolate and most relate genera
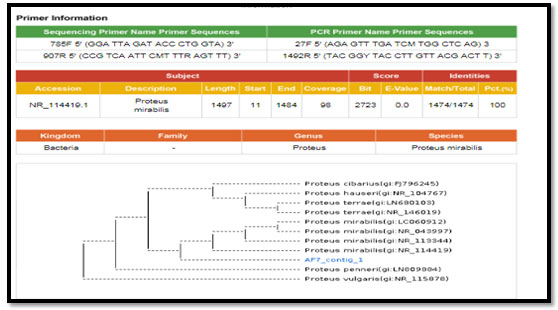
Figure 4: phylogenetic molecular tree of Pseudomonas spp. isolate and most relate genera
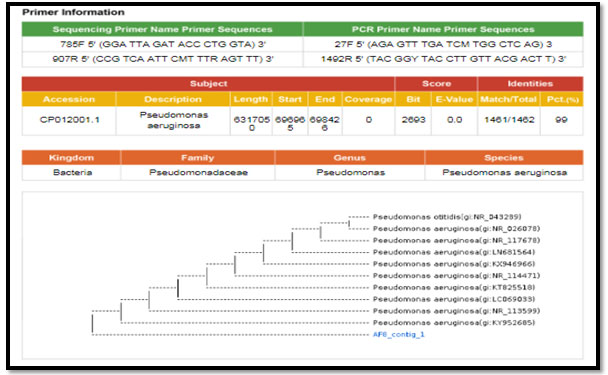
Figure 5: phylogenetic molecular tree of Staphylococcus lentus isolate and most relate genera
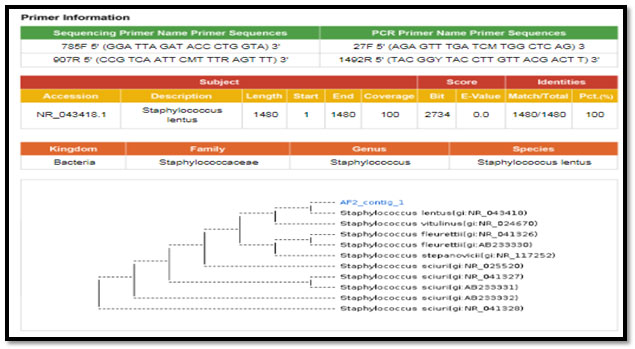
Figure 6: phylogenetic molecular tree of Staphylococcus sciuri isolate and most relate genera
Bacteriological profile of the minced chevon samples were as following; the positive aerobic bacteria were about 50/50 (100%) of the total minced chevon sample same result detected in case of Anaerobic, Enterobacteriacea spp. count while, only about 25/50 (50%) of tested minced chevon sample have Staphylococcus spp. Similar results of the total aerobic count was detected by (Ragab, et al. 2016; Shaltout, et al. 2016 and Mardziah, et al., 2019) during their assessment of the bacteriological quality of some meat products in the Egyptian retail markets. Also, nearly similar results observed by Bantawa, et al.
(2018) during their bacteriological evaluation of different meat samples of Dharan markets as; (54%) in case of staphylococcus sp. Lower results reported by Erdem, et al. (2014) during their microbiological quality survallaince in Istanbul as (96.67%). While, only about (24%) of Enterobacteracea sp. and (8%) of staphylococcus sp. was estimated by Salem, et al. (2018) in different meat sold in Menofia markets, Egypt. According to Ragab, et al. (2016) staphylococcus sp. was (20%). Aerobic Plate Count play an important role in judging of the hygienic conditions under which it has been produced, handled and stored as well as unsuitable condition during storage (Shaltout, et. al., 2016 and Zelalem, et al., 2019; Sial, et al., 2021).
Statistical analysis for Different Types of Micro-organisms (CFU/g) in the minced chevon samples were; the total aerobic counts reported about; 90×103, 38×104, 2.1×105 ± 2.1×104 as minimum, maximum, mean ±SE values respectively while, about; 16.2×104, 32.1×104, 2.1×105 ±1.2×10 CUF/g were detected as minimum, maximum, mean ±SE values respectively in case of total anaerobic count. Enterobacteriacea spp. showed about; 20×103, 6.0×105, 1.5×105 ±2.5×105 CUF/g as minimum, maximum, mean ±SE values respectively while, about; 10×103, 1.2×106, 1.6×105 ±3.7×105 CUF/g were detected as minimum, maximum, mean ±SE values respectively in case of Staphylococcus spp. counts. The Comparison with microbiological criteria for foodstuffs (GSO 1016/ 2015) viewed that; all tested chevon meat samples were within permissible limit and fit for human consumption. Nearly similar results found by Salem, et al. (2010) whome reocrded about 5.61×105CFU/g of total aerobic counts from grand total of thirty random different meat samples were collected from different butcher shops in Kaluobyia governorate, Egypt.
Higher result reported by Erdem, et al. (2014) were total aerobic counts were (9×106 CFU/g in minced meat) and by Ragab, et al. (2016) who detected about (6.6×108 CFU/g) of total aerobic counts in minced meat). Salem, et al. (2018) recorded about (7.35×104 CFU/g) of Enterobacteracea sp.in meat. While Tefera, et al. (2019) also recorded about (4.27×103 CUF/g) of Enterobacteracea sp. in minced meat. While about 5.6×105 CUF/g of staphylococcus sp. was detected by Haileselassie, et. al., (2013). Gonulalan and Kose, (2003) recorded about (6.7×106 CUF/g) Staphylococcus spp. from Turkey minced meat samples. Lower result observed by Shaltout, et al. (2016) who obtained about 8.03×104 CUF/g of total aerobic count while, in case of Enterobacteracea sp. they recorded about (2.02×102 CUF/g) and isolated about (2.67×102 CFU/g) of staphylococcus in Egyptian meat.
Hazaa, and El-Shater, (2019) who recoded about 1.21×103 staphylococcus sp. in meat. The results recorded by (Salem et al., 2018) in case of Enterobacteracea sp. was (7.35×104 CFU/g). while, in another study performed by Hassanien et al., (2018) about (4.27×103 CUF/g) of Enterobacteracea sp. was obtained. Enterobacteriaceae group consider one of the most challenging bacterial contaminants to meat globally. E. coli, Salmonella, klebsiella species and Proteus species, are the most common food poisoning that associated with meat (Al-Mutairi, 2011). The presence of Staphylococcus aureus a indicated improper hygienic practice and posed a risk to consumer safety (Abdelrahman et al., 2016).
Quick, sensitive, specific, and easy techniques for detection of the foodborne microorganisms needed for effective implementation of food safety. Polymerase chain reaction (PCR) became advent from 1980s and become one of the basic tool in molecular diagnostics and can be very efficiently used in rapid detection of food-borne pathogens (Armany et al., 2016). PCR identification mentioned in table (5) and figure (5) were as following; Staphylococcus sciuri were 2/9 (22.2), while; Aeromonas enteropelogenes, Escherichia fergusonii, Proteus mirabilis, Pseudomonas aeruginosa and Staphylococcus lentus were 1/9 (11.1) respectively.
Aeromonas is one of facultative anaerobic gram-negative, non-spore forming, rod-shaped, bacterium morphologically resembles members of the family Enterobacteriaceae. The most important pathogens are; A. enteropelogenes, A. caviae, A. hydrophila, and A. veronii biovarsobria. The organisms that are widely distributed in mainly among aquatic habitats. A. enteropelogenes is virulent pathogenic bacterium but its pathogenicity is still under investigation. This is the first report isolated of Aeromonas enteropelogenes from chevon meat (Aberoum & Jooyandeh 2010 and Silva, et al., 2019; Sial, et al., 2021).
Aeromonas spp. are pathogenic for man. Gastroenteritis, septicemia, muscle infections, soft-tissue and skin diseases are one of the most common illnesses caused by pathogenic Aeromonas spp. (Igbinosa et al. 2012). Aeromonas species has virulence activity on the cell structural including lipopolysaccharides (LPS), haemolysis, outer membrane proteins (OMPs), pili, flagella, toxins, that have a vital pathogenic role to the host (Matys, et al, 2020). Aeromonas enteropelogenes considered one of sever pathogenic bacterium (Ramesh and Souissi, 2018). The pathogenicity depends on the microbial hemolytic toxins which lysis of neutrophils and erythrocytes. A. enteropelogenes had β_ haemolytic action (Mogrovejo et al., 2020). The microorganisms mainly produce haemolysin which help them to adhere in the mucosal gut epithelial cells before starts its multiplication (Gudeta et al., 2016; Matys, et al, 2020; Sial, et al., 2021).
Escherichia fergusonii is a rod-shaped Gram-negative species, usually motile, and catalase positive, ferment D-glucose of bacterium, reduce nitrate to nitrite, they are positive for methyl red, acetate utilization, indole production & motility, and negative for the H2S production on triple sugar-iron agar, Voges-Proskauer reaction, urea hydrolysis, phenylalanine deaminase. This isolates highly related to; Escherichia coli, while E. fergusonii usually isolated from human blood samples. This microorganism considering as opportunistic pathogens of humans and were reported from human clinical specimens of an outbreak of food poisoning (Mogrovejo-Arias et al., 2020).
The study concluded that four types of Escherichia species were isolated from raw meat in Khartoum State, Sudan: E. coli, E. vulneris, E. albertii and E. fergusonii. were existed in meat samples. The presence of foodborne microorganisms including for example, E. coli samples reflects the role of meat as major reservoir for causative pathogenic agents (Ahmed and Al Sanosi, 2018). Similar results recorded by Mahapatra, and Mahapatra, (2005) who recognized that Escherchia fergusonii as a pathogen member of family Enterobacteriaceae. Escherichia fergusonii may be found in humans or animals as pathogens or commensals. On the other hand, E. coli considering one of the most public foodborne illnesses which has significant public health concern (Luna-Gierke et al., 2014).
Proteus mirabilis, is one of the Gram-negative Enterobacteriaceae family; facultative anaerobic, bacilli rod-shaped bacterium and resides in normal flora of man intestine. Proteus bacilli are widely distributed in nature as saprophytes, about one from each four persons of the population suffering from P. mirabilis in their fecal matters in addition to animal matter, sewage, manure soil, the mammalian intestine and animal feces. This opportunistic nosocomial pathogen may cause urinary septic infections. Proteus mirabilis causes 90% of humans Proteus infections. Pathogenicity of P. mirabilis pathogenesis by two steps; firstly, by colonization of the microorganisms in the urinary tract followed by complete evade of the body defense (Schaffer & Pearson, 2017 and Armbruster et al., 2018; Mogrovejo-Arias et al., 2020; Milton et al., 2021).
mirabilis is one of the seldom food borne microorganisms which transmitted from seafood, vegetables, and meat (Wang et al., 2019). P. mirabilis reported one of the most food poisoning microorganism in China. The clinical symptoms of P. mirabilisinfection including; fever, dizziness, abdominal pain, nausea, diarrhea and vomiting after 0.67–9 h incubation period (Huo et al., 2014). About 3.61% P. mirabilisfood poisoning incidents recorded in Datong from 2016 to 2017 (Shanxi Province, China) (Gong et al., 2019).
P. mirabilis play an important role in food spoilage and considering as enteropathogens (Kushwaha et al., 2014). Pseudomonadaceae family containing 191 species, Pseudomonas is gram-negative, encapsulated, rod-shaped bacterium. P. aeruginosa is considered all over the world as one of the most dangerous organisms causing different diseases and capable of secreting many extra cellular products which play a role in the virulence of pathogenic strains of P. aeruginosa (Pang et al., 2019; Milton et al., 2021)
aeruginosa may infect animal, plant, and commonly be opportunistic to the human as it mainly affecting the immunocompromised persons through cystic fibrosis or through burned tissues and traumatic tissues (Bassetti, et al., 2018). P. aeruginosa has antibiotic resistance, and considered nosocomial infection including various sepsis syndromes and ventilator-associated pneumonia (Ruffin, M. and Brochiero, 2019). P. aeruginosa infections hardly treated due to its natural antibiotics’ resistance. P. aeruginosa is present in the intestinal tract of both man and animals and its presence in food could be taken as an index of fecal contamination (Mostafa et al., 2018).
These pathogenic strains play an important role in bloodstream infection and respiratory tract infections, mastitis, endometritis, chronic pulmonary disease, urogenital tract infection, cystic fibrosis and sever form of gastroenteritis among man, animals and sometimes may cause fatal infections specially with the immunodeficient persons (Rocha, et al., 2019). Pseudomonas aeruginosa was detected in many food items. Although it did not identify as one of food poisoning microorganisms, but it mainly spoil food. The infection mainly transmitted through food sold in open air which exposed to dust and flies (Alayande, et al., 2018). Staphylococcus aureus cause food intoxication (Chikwanha, et al., 2018). Staphylococcus lentus is oxidase-positive, coagulase-negative, gram positive member of Staphylococcus genus.
These microorganisms are related originally to Staphylococcus sciuri derived from subspecies “lentus” (Shaker et al., 2018). Staphylococcus lentus is colonize on the skin of human and animals and reported as commensal bacterium. It has commonly isolated from food-producing animals, including dairy animals, poultry, and their food products. Animals man workers recorded as carriers of S. lentus (Schwendener and Perreten, 2012). Staphylococcus lentus forming biofilm that resist antibiotics which increase mortality rate as a result of the difficulty to controlling the infections (Al-Azawi et al., 2018; Mogrovejo-Arias et al., 2020; Milton et al., 2021).
Consumption of foods contaminated Staphylococcus lentus have been described as able to produce enterotoxins (Zabrodskii, 2020; Milton et al., 2021). Staphylococcus sciuri is kwon as animal-associated microorganisms in addition to its presence on mucosal and skin surfaces of farm, wild animals, and pets and in animal origin food items, its clinical importance for man is increasing. Staphylococcus sciuri is novobiocin-resistant, oxidase-positive, coagulase-negative staphylococcal species. It is widely distributed in environmental as reservoirs including water, soil, sand, and marsh grass (Lu, et al., 2020).
S. sciuri is widely found in environment and from several animals and animals’ products (Heilmann et al., 2019) as well as from human, this microorganism considers as animals’ pathogens (Koli et al., 2018). Their signs containing; septic shock, endocarditis, pelvic inflammation, peritonitis, endophthalmitis, and wound infections and urinary tract infection (Kentzi et al., 2016). S. sciuri may causing ruminants mastitis especially in goats and cow. There was a big shortage on information about S. sciuri pathogenicity in animals (Romanò et al., 2020; Milton et al., 2021).
CONCLUSION
Chevon can be considered best replacement of beef meat due to its lower unhygienic total fat; “cholesterol and saturated fat” and its lower calories content in comparison to beef content, which protect the consumers health. All tested chevon meat samples were within permissible limit and fit for human consumption. Molecular Identification of isolated microorganism declared the following: Staphylococcus sciuri, Aeromonas enteropelogenes, Escherichia fergusonii, Proteus mirabilis, Pseudomonas aeruginosa and Staphylococcus lentus. This is the first report isolated of Aeromonas enteropelogenes from chevon meat. All isolated microorganisms had public health concern as food poisoning microorganisms. Further investigation needed to study the chevon meat and its microbial quality.
Financial Support: The article is self-funded by the authors.
Conflict of Interest: Authors declares no conflicts of interests to disclose.
Ethical Clearance Statement: The Current Research Work Was Ethically Approved by the Institutional Review Board (IRB) of College of Science, University of Jeddah Saudi Arabia.
REFERENCES
Abdelrahman, H., Ismail, S. & Harydi, A. (2016).Assessment of the Bacteriological Quality of Minced Meat and Beef Burger at Selected Egyptian Hypermarkets. Suez Canal Veterinary Medicine Journal. SCVMJ Vol 21(1), pp 91-97.
Abebe, E., Gugsa, G., Ahmed, M. (2020). Review on Major Food-Borne Zoonotic Bacterial Pathogens. Journal of Tropical Medicine. Pp 1-19.
Aberoum, A. & Jooyandeh, H. (2010). A review on occurrence and characterization of the Aeromonas species from marine fishes. World J Fish and Marine Sci Vol 2, pp 519– 523.
Ahmed, A., & Al Sanosi, M. (2018). Occurrence of Escherichia species in vended red meat in Khartoum State, Sudan. Journal of Veterinary Medicine and Animal Production, Vol 8(1) pp 2-12.
Alayande AB, Aung MM, Kim IS. (2018). Correlation Between Quorum Sensing Signal Molecules and Pseudomonas aeruginosa’s Biofilm Development and Virulency, Current Microbiology Vol 75, pp 787-793
Al-Azawi, I. H., Al-Hamadani, A. H., & Hasson, S. O. (2018). Association between biofilm formation and susceptibility to antibiotics in Staphylococcus lentus isolated from urinary catheterized patients. Nano Biomed. Eng, Vol 10(2), pp 97-103.
Al-Mutairi, M. F. (2011). The incidence of Enterobacteriaceae causing food poisoning in some meat products. Advance Journal of Food Science and Technology Vol 3(2), pp 116-121.
American Public Health Association (2002). Methods for the Microbiological Examination of Foods 4th Ed., APHA Technical Committee on Microbiological Methods for Foods. Washington, D.C., USA.
Armany, G., Ibrahim, H., Amin, R. & Ahmed, H. (2016). Detection of some foodborne pathogens in meat products by Polymerase Chain Reaction. Benha Veterinary Medical Journal Vol 30(1), pp 323-330.
Armbruster, C. E., Mobley, H. L., & Pearson, M. M. (2018). Pathogenesis of Proteus mirabilis infection. EcoSal Plus, Vol 8(1), pp 1-13.
Aseel, A. S., Mayada, F. H., & Majed, H. M. (2011). Isolation and molecular identification of Salmonella typhimurium from chicken meat in Iraq. J World’s Poult Res, Vol 3(2), pp 63-66.
Bantawa, K., Rai, K., Limbu, D. & Khanal, H. (2018). Food-borne bacterial pathogens in marketed raw meat of Dharan, eastern Nepal. BMC research notes Vol 11(1), pp 1-5.
Bassetti, M., Vena, A., Croxatto, A., Righi, E., and Guery, B. (2018). How to manage Pseudomonas aeruginosa infections. Drugs Context Vol 7, pp 21-34.
Chikwanha, O.C.; Vahmani, P.; Dugan, M.E.R.; Mapiye, C. (2018). Nutritional enhancement of sheep meat fatty acid profile for human health and wellbeing. Food Res. Int, Vol 104, pp 25–38.
Erdem, K., Saglam, D., Ozer, D. and Ozcelk, E. (2014). Microbiological quality of minced meat samples marketed in Istanbul. Van Veterinary Journal Vol 25(3), pp 67-70.
Figueras, M. J., Beaz‐Hidalgo, R., Senderovich, Y., Laviad, S., & Halpern, M. (2011). Re‐identification of Aeromonas isolates from chironomid egg masses as the potential pathogenic bacteria Aeromonas aquariorum. Environmental Microbiology Reports, Vol 3(2), pp 239-244.
GCC Standrization Organization (GSO) 1016/2015. Microbiological Critieria for Foodstuffs. Meat and poultry and their products. Pp 12-14.
Gecgel, S. K., & Demircan, N. (2016). The epidemiology of pathogen microorganisms in hospital acquired infections. Int J Clin Exp Med, Vol 9(11), pp 22310-22316.
Gong, Z., Shi, X., Bai, F., He, X., Zhang, H., YuBin, L., & Hu, Q. (2019). Characterization of a novel diarrheagenic strain of Proteus mirabilis associated with food poisoning in China. Frontiers in Microbiology, Vol 10, pp 10-28.
Gonulalan, Z. & Kose, A. (2003). Microbiological quality of ground beef retailed in Kayseri. Fırat Univ. Sağ. Bil. Vet. Derg, Vol 17, pp 49-53.
Gudeta, D. D., Bortolaia, V., Amos, G., Wellington, E. M. H., Brandt, K. K. and Poirel, L. (2016). The soil microbiota harbors a diversity of carbapenem-hydrolyzing β-lactamases of potential clinical relevance. Antimicrob. Agents Chemother. Vol 60, pp 151–161.
Haileselassie, M., Taddele, H., Adhana, K., & Kalayou, S. (2013). Food safety knowledge and practices of abattoir and butchery shops and the microbial profile of meat in Mekelle City, Ethiopia. Asian Pacific journal of tropical biomedicine, Vol 3(5), pp 407-412.
Hassanien, F., Shaltout, F., Mohamed H. & Hatem, E. (2018). Quality assurance of some locally processed meat products. Benha Veterinary Medical Journal Vol 34(1), pp 41-47.
Hauschild, T., & Schwarz, S. (2003). Differentiation of Staphylococcus sciuri Strains Isolated from Free‐Living Rodents and Insectivores. Journal of Veterinary Medicine, Series B, Vol 50(5), pp 241-246.
Hazaa, W., & El-Shater, M. (2019). Identification of Some Biological Hazards in Some Meat Products. Benha Veterinary Medical Journal, Vol 37(1), pp 27-31.
Heilmann C, Ziebuhr W, Becker K. (2019). Are coagulase-negative staphylococci virulent? Clin Microbiol Infect. Vol 25(9), pp 1071–1080.
Huo, Z., Bai, S. Y., Gao, B., & San-Mei, H. U. (2014). A study on the food poisoning isolated Vibrio parahaemolyticus and Proteus mirabilis. Chin. J. Health Lab. Technol. Vol 24, pp 3254–3256.
Igbinosa, I.H., Igumbor, E.U., Aghdasi, F., Tom, M. & Okoh, A.I. (2012). Emerging Aeromonas species infections and their significance in public health. Sci World J 2012, 625023.
ISO 21528-2 (2004). Microbiology of food and animal feeding stuffs – Horizontal methods for the detection and enumeration of Enterobacteriaceae – Part 2: Colony-count method
ISO 6888-2 (1999). Microbiology of food and animal feeding stuffs – Horizontal method for the enumeration of coagulase-positive staphylococci (Staphylococcus aureus and other species) – Part 2: Technique using rabbit plasma fibrinogen agar medium.
Janda, J. M., & Abbott, S. L. (2010). The genus Aeromonas: taxonomy, pathogenicity, and infection. Clinical microbiology reviews, Vol 23(1), pp 35-73.
Jiang, J., & Xiong, Y. L. (2016). Natural antioxidants as food and feed additives to promote health benefits and quality of meat products: A review. Meat science, Vol 120, pp 107-117.
Kentzi D, Kolyva S, Spiliopoulou I, Marangos M, Dimitriou G. (2016). Treatmentoptions for persistent coagulase negative Staphylococcal bacteremia in neonates. Curr Pediatr Rev. Vol 12(3), pp 199–208.
Koli CE, Njoga EO, Enem SI, Godwin EE, Nwanta JA, Chah KF (2018). Prevalence, toxigenic potential, and antimicrobial susceptibility profile of Staphylococcus isolated from ready-to-eat meats. Vet World. Vol 11(9), pp 1214–1221.
Kushwaha, K., Babu, D., & Juneja, V. K. (2014). Proteus, in Encyclopedia of Food Microbiology (Second Edition), eds C. A. Batt and M. L. Tortorello, (Oxford: Academic Press), pp 238–243.
Lu, Y.; Lu, Q.; Cheng, Y.; Wen, G.; Luo, Q.; Shao, H. and Zhang, T. (2020). High concentration of coagulase-negative staphylococci carriage among bioaerosols of henhouses in Central China. BMC Microbiology. Vol 20, pp 21-30.
Luna-Gierke, R. E., Griffin, P. M., Gould, L. H., Herman, K., Bopp, C. A., Strockbine, N., & Mody, R. K. (2014). Outbreaks of non-O157 Shiga toxin-producing Escherichia coli infection: USA. Epidemiology & Infection, Vol 142(11), pp 2270-2280.
Mahapatra, A., & Mahapatra, S. (2005). Escherichia fergusonii: an emerging pathogen in South Orissa. Indian Journal of Medical Microbiology, Vol 23(3), pp 204.
Mardziah, A., Yeo, M., Puah, H., Chua, H., Chew, K. (2019). Staphylococcus aureus infections in Malaysia: a review of antimicrobial resistance and characteristics of the clinical isolates, 1990–2017, Antibiotics, Vol 8, pp 1–29.
Matys, J.; Turska-Szewczuk, A. and Sroka-Bartnicka, A. (2020). Review: Role of bacterial secretion systems and effector proteins – insights into Aeromonas pathogenicity mechanisms. ABP Acta Biochimica Polonica. Vol 67(3), pp 283-293.
Mazhangara, I.R.; Chivandi, E.; Mupangwa, J.F. and Muchenje, V. (2019). The Potential of Goat Meat in the Red Meat Industry. Sustainability Vol 11(13), pp 3671-3683.
Milton, A.A.P., Momin, K.M., Priya, G.B., Ghatak, S., Das, S., Gandhale, P.N., Angappan, M. and Sen, A., (2021). Development of novel visual detection methodology for Salmonella in meat using saltatory rolling circle amplification. Journal of Applied Microbiology.
Mogrovejo, D.C.; Perini, L.; Gostinčar, C.; Turk, M.; Ambrožič-Avguštin, J.; Brill, F. H. and Gunde-Cimerman, N. (2020). Prevalence of Antimicrobial Resistance and Hemolytic Phenotypes in Culturable Arctic Bacteria. Front. Microbiol. Vol 13(5), pp 50-59.
Mogrovejo-Arias, D. C., Brill, F. H. H., and Wagner, D. (2020). Potentially pathogenic bacteria isolated from diverse habitats in Spitsbergen, Svalbard. Environ. Earth Sci. pp 79:109.
Mostafa, A. A., Al-Askar, A. A., Almaary, K. S., Dawoud, T. M., Sholkamy, E. N., & Bakri, M. M. (2018). Antimicrobial activity of some plant extracts against bacterial strains causing food poisoning diseases. Saudi Journal of Biological Sciences, Vol 25(2), pp 361-366.
Pal, M., Ayele, Y., Patel, A. S., & Dulo, F. (2018). Microbiological and hygienic quality of Meat and Meat Products. Beverage and Food World, Vol 45(22), pp 21-7.
Pang, Z.; Raudonis, R.; Glick, B.; Julin, T. and Cheng, Z. (2019). Antibiotic resistance in Pseudomonas aeruginosa: mechanisms and alternative therapeutic strategies. Biotechnology Advances. Vol 37(1), pp 177-192.
Priya, G.B., Agrawal, R.K., Milton, A.A.P., Mendiratta, S.K., Singh, B.R., Kumar, D., Mishra, M. and Gandham, R.K., (2021). Isothermal amplification assay for visual on-site detection of Staphylococcus aureus in Chevon. Food Biotechnology, pp.1-16.
Ragab, W. S., Hassan, E.A., AL-Geddawy, M. A., & Albie, A.A. (2016). Bacteriological Quality of some Meat Products in the Egyptian Retail Markets. Assiut J. Agric. Sci. Vol 47, pp 422-429.
Ramesh, D. & Souissi, S. (2018). Antibiotic resistance and virulence traits of bacterial pathogens from infected freshwater fish, Labeo rohita. Microbial pathogenesis, Vol 116, pp 113-119.
Rocha, A.J.; Barsottini, M.R.; Rocha, R.R.; Laurindo, M.V.; de Moraes, F.L. and Rocha, S.L. (2019). Review: Human & Animal Health Pseudomonas aeruginosa: Virulence Factors and Antibiotic Resistance Genes. Brazilian Archives of Biology and Technology. Vol 62, pp 19-29.
Romanò, A.; Gazzola, A.; Bianchini, V.; Cortimiglia, C.; Maisano, A.; Cremonesi, P.; Graber, H.; Vezzoli, F. and Luini, M. (2020). Staphylococcus aureus From Goats Are Genetically Heterogeneous and Distinct to Bovine Ones. Front Vet Sci. Vol 7, pp 628-637.
Ruffin, M. and Brochiero, E. (2019). Repair Process Impairment by Pseudomonas aeruginosa in Epithelial Tissues: Major Features and Potential Therapeutic Avenues. Front. Cell. Infect. Microbiol. Vol 9, pp 182-200.
Salem, A., Amin, A. & Afifi, S. (2010). Studies on Antimicrobial and Antioxidant Efficiency of Some Essential Oils in Minced Beef. J. American Sci. Vol 6, 691-700.
Salem, A., Shawky, N. & Abo-Hussein, L. (2018). Microbiological Profile of Some Meat Products in Menofia Markets. Benha Veterinary Medical Journal Vol 34(2), pp 1-7.
Schaffer, J. N., & Pearson, M. M. (2017). Proteus mirabilis and urinary tract infections. Urinary Tract Infections: Molecular Pathogenesis and Clinical Management, pp 383-433.
Schwendener, S., & Perreten, V. (2012). New MLSB resistance gene erm (43) in Staphylococcus lentus. Antimicrobial agents and chemotherapy, Vol 56(9), pp 4746-4752.
Shaker, M. N., Hmdan, T. A., & Issa, A. H. (2018). Isolation and diagnosis of Staphylococcus lentus from different operation theater hospitals. Sci J Med Res, Vol 2(8), pp 177-181.
Shaltout, F. A., Salem, A. Khater, D. & Lela, R. (2016). Impact of some natural preservatives on Bacterial Profile of Minced Meat in Egypt. Benha Veterinary Medical Journal Vol 31(1), pp 35-42. Sial, F., Barham, S., Shah, A., KhaSKheli, G., Jamali, M., Siyal, P. (2021). Physico-Chemical Quality and Calorific Value of Buffen, Venison and Chevon. Journal of Animal Health and Production. Vol 9(2), pp 178-184.
Silva, A., Barros, L., Virgens, D., Velame, D. (2019). Aeromonas spp in fish and in continental waters. Revista Brasileira de Higiene e Sanidade Animal Vol 13(1), pp 15 – 47.
Singh, P. K., Kumar, S., Kumar, P., & Bhat, Z. F. (2014). Effect of mincing on the quality characteristics of chevon cutlets. Journal of Animal Research, Vol 4(2), pp 193-200
Spilker, T., Coenye, T., Vandamme, P., & LiPuma, J. J. (2004). PCR-based assay for differentiation of Pseudomonas aeruginosa from other Pseudomonas species recovered from cystic fibrosis patients. Journal of clinical microbiology, Vol 42(5), pp 2074-2079.
Tefera, M., Aleme, S., Girma, H. (2019). Antimicrobial sus-ceptibility pattern of S. aureus isolated from sheep and goatcarcasses, Open Microbiology Journal, Vol 13(1), pp 16–20.
Vita Lele, S., Sidlauskiene, D., Klupsaite, V., Buckiuniene, E., Viskontaite, J., Klementaviciute, P., Zavistanaviciute, V., Sakiene, E., Bartkiene, A. (2020). Comparison studies of the chemical, physical, technological, and microbiological characteristics of the European roe deer, boar, red deer, and beaver hunted wild game meat. Japan. Soci. Anim. Sci., pp 133-146.
Wang, S., Xu, L., Chi, X., Li, Y., Kou, Z., Hou, P. (2019). Emergence of NDM-1- and CTX-M-3- producing Raoultella ornithinolytica in human gut microbiota bacterial isolation and identification. Front. Microbiol. 10:2678.
Zabrodskii, P. F. (2020). Food Poisoning. Bacteria Associated with Food. ACTA Scientific Microbiology. Vol 3(3), pp 1-10.
Zelalem, M., Sisay, L., Vipham, K., Abegaz, A., Kebede, B. and Terefe, Y. (2019). prevalence and antimicrobial resistance profiles of bacterial isolates from meat and meat products in Ethiopia: a systematic review and meta-analysis, International Journal of Food Contamination, Vol 6(1), pp 1–14.
Zlatian O, Balasoiu AT, Balasoiu M, Cristea O, Docea AO, Mitrut R, SpandidosDA, Tsatsakis AM, Bancescu G, Calina D. (2018). Antimicrobial resistance in bacterial pathogens among hospitalized patients with severe invasive infections. ExpTher Med. Vol 16(6), pp 4499–510.


