1 Laboratory of Plant Molecular Genetics and Genomics, Institute of Bioresources & Sustainable
Development, Takyelpat, Imphal 795001, Manipur, India
2 Department of Biotechnology, Gauhati University, Guwahati 781014, Assam, India
Corresponding author email: premi.pukhrambam@gmail.com
Article Publishing History
Received: 17/03/2023
Accepted After Revision: 24/03/2023
Polygonum is an important genus of Polygonaceae and Polygonum posumbu is underutilized traditional medicinal plant whose leaves are extensively used as an antipyretic and dyspepsia agent in Manipur, India as well as a spice in India, Japan, Nepal, China, South Korea, Philippines and Thailand. P. posumbu is commonly known as Phak-pai in Manipur. DNA barcode based molecular characterization technology for species identification has been recognized as a reliable tool for plants but the selection of suitable universal marker is still under discussion.
The main aim of the study is to identify the Polygonum posumbu sample from Manipur, India using DNA barcode method. In the present study, Polygonum posumbu samples were collected from a local farmer in Awang khunou, Imphal West, Manipur, India which was taxonomically identified. To assess the molecular species identification, Internal Transcribed spacer (ITS) marker system was employed and the primer’s pair used were covered the internal transcribed spacer 1, 5.8S ribosomal RNA gene and internal transcribed spacer 2 complete sequence. Phylogenetic tree was constructed for the studied sample with the sequences of the 9 species retrieved from the GenBank in order to show the relationships among the samples. The result shows that the ITS marker-based DNA identification method successfully characterized the Polygonum posumbu species. The information of ITS marker-based identification of Polygonum posumbu from Manipur, India, will enhance our knowledge in better understanding the medicinal properties of this plant.
DNA Barcode, Identification, ITS Marker, Phylogenetic Tree and Polygonum posumbu.
Pukhrambam P. D, Devi K. K, Das S. Molecular Identification of Medicinal Plant, Polygonum posumbu from Manipur, India. Biosc.Biotech.Res.Comm. 2023;16(1).
Pukhrambam P. D, Devi K. K, Das S. Molecular Identification of Medicinal Plant, Polygonum posumbu from Manipur, India. Biosc.Biotech.Res.Comm. 2023;16(1). Available from: <a href=”https://bit.ly/3lGPfGr“>https://bit.ly/3lGPfGr</a>
INTRODUCTION
According to reports, North-East India has used more than 2000 kinds of ethnobotanical plants to make a variety of remedies (Schori and Showalter 2011). Polygonum posumbu Buch.-Ham.ex D. Don, a small herb, which is distributed in the North-Eastern states of India. Manipur is one of the North-Eastern states and here P.posumbu is used as an ethnomedicinal plant in various ways. Traditionally, tender shoots of Polygonum posumbu is consumed as a spice and leaves are used as mouth-freshener (Asha et al. 2011). As a folk medicine, it is used for curing dyspepsia and fever (Singh et al. 2003; Yumnam et al. 2012). The juice of the tender shoots and fresh leaves are used to reduce heartbeat (Leishangthem and Dinendra 2014 Reshmi Singh et al 2016, Mosa et al 2019 , Cahyaningsih et al 2022).
This plant also possesses antibacterial activity against Clostridium sporogens, Neiserria gonorrhea and Bacillus cereus (Ishwori et al. 2014). A great source of unidentified chemical compounds with therapeutic benefits is medicinal plants (Rao 2004). There is an urgent need for precise taxonomy inventorization in relation to species-level genetic characterization of the plant from this region, as the majority of the people in this place used the traditional knowledge of herbal medicine. Despite the medicinal value of Polygonum posumbu in traditional medicinal system, this plant does not have any other information including pharmaceutical, genetics and genomics information.
The main concerns in ethnobotany focused on the significance of accurate species identification as well as the interpretation of indigenous and conventional knowledge of restorative plant usage and their transfer for the advancement of bioprospect in human health care. Currently, the re-evaluation of traditional medicine is being done all over the world via comprehensive research on various plant species and their therapeutic properties. Consumer demand is currently encouraging the expansion of dietary supplements and innovative herbal plant-based treatments. In the healthcare sector of the 21st century, use of herbal plant based dietary supplements or medication is rapidly growing (Siew et al. 2014, Cahyaningsih et al 2022).
Due to misidentification, there is the probability of substitution within certain plants which may give different affects rather than the expected certain properties from that of the original plant. Therefore, proper identification of herbal medicinal plants in relation to their naturalness and lack of adulteration and safe application has become increasingly important (Pang and Chen 2014).An ideal strategy for the preservation of plant genetic resources and genetic advancement is the characterization of plants using morphological and molecular markers (Rout and Mohapara 2008). Both molecular and morphological characteristics are important for the identification of underutilized medicinal plants and the combined data would provide more comprehensive information for those plants.
The appraisal and exploitation of genetic resources can also benefit from the use of morphological and molecular data. Plant genetic diversity has been described and evaluated using morphological characteristics. However, morphological characteristics, particularly quantitative traits are not stable as environmental conditions from one region to other region may have impacts on the morphology of the plant. The majority of the current taxonomy plant group identification is morphological in nature. Nevertheless, this identification technology has several limitations when attempting to distinguish between plants at different developmental stages. Since molecular markers are not affected by environmental factors, they are a useful complement (Solmaz et al. 2010 , Cahyaningsih et al 2022).
DNA barcoding has become a highly effective method for identifying species, as seen by the widespread use of this method in bioresource monitoring and documentation (Kress et al. 2005; Hollingsworth et al. 2011; Stoeckle et al. 2011 and Liu et al. 2012, Cahyaningsih et al 2022). The technique’s application stresses some key areas, such as documenting the significant and fragile ethnomedicinal plant bioresources, with which the field of “Ethnobotany Genomics” has just been created (Newmaster et al. 2010). The purpose of DNA barcoding is clearly to identify an unknown sample using pre-existing classification but it will not detect relationship between samples. The basic goal of DNA barcoding is to identify a universal DNA sequence that balances conserved sequences with enough variability to distinguish across different organisms. The basic materials used to create herbal goods can be identified using barcoding technology. The “Consortium for the barcode of life plant working group (CBOL)” has accessed numerous chloroplast genomic areas (ITS, 5S rRNA, rbcL, rpl36-rps8, matK and trnH-psbA) in plant systems (CBOL 2009).
Among the chloroplast genomic areas used for plant identification, internal transcribed spacer (ITS) or its segments (ITS1, ITS2) is one of the most frequently used regions in plant molecular systematic at generic and species levels. This is due to the fact that, ITS has the potential to provide a clear differentiation of relationships between and within species (Hillis and Dixon 1991; Yuan et al. 2015; Buchheim et al. 2011; Alvarez and Wendel 2003; Staggermeier et al. 2015; Keller et al. 2010 and Baldwin et al. 1995). It was used as a seed plant barcode (Song et al. 2012; Hollingsworth 2011 and Li et al. 2011). Studies showed that, species identification using barcodes from both the uniparentally inherited plastid genome and biparentally inherited nuclear genome is more accurate or even required and the nuclear genome’s ITS region is now the most promising candidate (Fazekas et al. 2009; Chase and Fay 2009 and Roy et al. 2010).
An example for the use of DNA barcoding genes for identification of species is the illustration of the discrimination of Schisandra chinensis at the species and population levels with the use of ITS2 (Li et al. 2013). Additionally, S. Chinensis from S. sphenanthera could be distinguished by their ITS2 region with great clarity. In a study to identify Astragalus 319 species were examined across four coding (trnH-psbA, rpoC1, rbcL and matK) and two non-coding (ITS and ITS2) areas; ITS2 and ITS barcodes were more effective at differentiating the species (Gao et al. 2009).
The situation where Peucedanum praeruptorum has been distinguished by DNA barcoding. In order to identify it from typical knockoffs and adulterants, Zhou et al. 2014 employed ITS and nrDNA barcodes. Despite the fact that, the Polygonum posumbu has high traditional economic importance, owing to their medicinal and nutritional value, till date, there is no information concerning chemical constituents as well as genetic information, and pharmacologic activity of the plant. Therefore, the main purpose of our study was to identify Polygonum posumbu plant commonly found in Manipur, India. The data will be helpful in further exploration of Polygonum posumbu and also in functional genomics for improving the medicinal uses of P. Posumbu.
MATERIAL AND METHODS
Sample collection: For the study, healthy Polygonum posumbu plants were collected from a local farmer in Awang khunou, Imphal West, Manipur, India (N 24°47.177’ E 093°50.981’) in November 2017. Leaf tissues of plant samples were cleaned, and then stored in a sterile polyethylene bag at -80°C until further analysis. For morphological identification herbarium was prepared using 4% formalin. The plant sample was identified based on morphological and taxonomical characteristics by Dr. B. Thongam, Taxonomist, IBSD, Manipur, India. A voucher specimen was deposited in the herbarium of the Institute of Bioresources and Sustainable Development (IBSD), Manipur, India. The plant sample was also identified by the Botanical Survey of India, Eastern Regional Centre (BSI ERC), Shillong.
DNA isolation:Fresh plant leaves were washed with sterile distilled water and scraped. Approximately 100mg of leaves were sliced into thin pieces and ground to a fine paste using liquid N2. Total genomic DNA was extracted by using DNeasy Plant Mini Kit (Qiagen). The isolated DNA was purified and quantified in gel electrophoresis using gel red stained band intensity and concentration and purity of DNA were checked in Bio Spec Nano (Shimadzu).
DNA amplification: The polymerase chain reaction (PCR) was performed using ITS-An5, 5´- CCTTATCATTTAGAGGAAGGAG – 3´, ITS-An4, 5´- CCGCTTATTGATATGCTTAAA – 3´ primer pair (Table 1). The PCR reaction of 25µl mixture consisted of 10X PCR reaction buffer (2.5 µl), 25mM MgCl2 (1.5 µl), 10 mM dNTPs (0.5 µl), 5 unit of Taq polymerase (0.25 µl), 10µM of each primer (1 µl),50ng genomic DNA (1 µl) and 20.25µl Milli-Q water (17.25 µl). PCR thermal conditions (on a C1000 Touch Thermal Cycler; Bio-rad) were 95°C for 3 min, 32 X [95°C for 30s, 52°C for 1 min, 72°C for 1min] and final extension at 72°C for 5min. The PCR product was checked by 1.5% agarose gel electrophoresis.
Purification of PCR products and DNA sequencing: Qiagen DNA Mini Kit (Germany) was used to purify the PCR product to ensure that the final products were free of contaminants. Purified PCR products were sent for Sanger sequencing at Europhins Genomics India Pvt., Ltd., Bangalore.
Sequence analysis: Utilizing BLASTn analysis, the ITS amplified sequence was BLAST in GenBank. The forward and reverse sequences were then aligned to the target sequences and raw sequences were edited manually. The aligned sequences were corrected manually and checked the nucleotide composition. For a sequence length of ̴ 724bp, the terminals 3´ and 5´ were cut to provide consensus sequence for the taxa. The modified sequence for the studied species was submitted to NCBI databases (http://www.submit.ncbi.nlm.nih.gov). Also, 9 sequences for the same or related species as the examined material were retrieved from GenBank (http://www.ncbi.nlm.nih.gov/genbank).
Phylogenetic analysis: The ITS region of 9 taxa from the Polygonaceae family was retrieved from GenBank for the phylogenetic tree construction in which 8 taxa are from same genus Persicaria and the 9th one is from the genus Antigonon (name of the species is Antigonon leptopus). The phylogenetic tree was constructed for the studied species and UPGMA analysis was done in Mega 11 software to examine the relationship between the studied species and 9 taxa retrieved from GenBank and bootstrap value of 1000 replicates was calculated.
RESULTS AND DISCUSSION
In this study, fresh plant sample of Polygonum posumbu collected from Manipur, India was characterized based on morphological traits. The voucher specimen number (IBSD/M-251) (fig. 1) and (No: BSI/ERC/Tech/2021/111) were assigned respectively from the Institute of Bioresources and Sustainable Development (IBSD), Manipur, India and Botanical Survey of India (BSI), Eastern Regional Centre (ERC), Shillong respectively.
Table 1. Sequence of primers used for molecular identification
| S. No. | Primer code | primer sequence | Nucleotide length (bp) |
| 1 | ITS-An5 | CCTTATCATTTAGAGGAAGGAG | 22 |
| 2 | ITS-An4 | CCGCTTATTGATATGCTTAAA | 21 |
Figure 1: Taxonomic identification of Polygonum posumbu in IBSD, Imphal

Species identification:Polygonum posumbu has been identified using ITS primer pair ITS-An5 and ITS-An4. In this investigation, the amplification of the primers was satisfactory and reproducible. Due to length variation in the ITS forward and reverse sequences, only 724 aligned nucleotide positions were used for sequence analysis. According to BLASTn analysis, the sample was accurately identified upto the species level proving that the preliminary identification of plant based on morphological and taxonomical characters matched with the scientific name received from GenBank. BLASTn analysis result for the sample investigated shows a maximum identity of 98.90% and query coverage of 100% which is showing that ITS marker-based identification of Polygonum posumbu obtained from Manipur is accurate (Table 2). The accession number of the sequence submitted to the NCBI databases is OP602225.
Table 2. BLAST analysis result of ITS sequence of Polygonum posumbu (studied sample)
| Sample sent to NCBI | Maximum identity | Query coverage | Sequence length | Accession No. |
| Persicaria posumbu Voucher BSI/ERC/Tech/2021/111 | 98.90% | 100% | 724 bp | OP602225 |
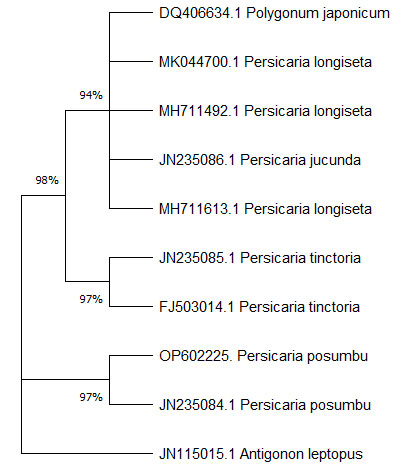
Phylogenetic analysis:UPGMA tree construction methods were used to construct dendrogram for the sequence of the studied sample and 8 sequences from same taxa and 1 sequence from different taxa (Table 3). The dendrogram derived from UPGMA analysis of the Maximum composite likelihood of all the 9 sequences with the sequence of the studied sample showed that the studied sample collected from Manipur, India (OP602225, Persicaria posumbu) is clustering with the sequences of the same taxa forming their own grouping and other 8 sequences are clustered away from the studied sample (Fig. 2).
Table 3. ITS sequences of Polygonaceae family retrieved from NCBI
| Species | Genus | Accession No. of ITS |
| Persicaria posumbu | Persicaria | OP602225 (studied sample) |
| Persicaria posumbu | Persicaria | JN235084.1 |
| Polygonum japonicum | Polygonum | DQ406634 |
| Persicaria jucunda | Persicaria | JN235086 |
| Persicaria longiseta | Persicaria | MH711613 |
| Persicaria longiseta | Persicaria | MH711492 |
| Persicaria longiseta | Persicaria | MK044700 |
| Persicaria tinctoria | Persicaria | JN235085.1 |
| Persicaria tinctoria |
Persicaria | FJ503014.1 |
| Antigonon leptopus |
Antigonon | JN115015.1 |
Figure:2 ITS sequences of Polygonaceae. A total of
In the present study, morphological identification was carried out first in order to identify the Polygonum posumbu collected from Manipur, India. However, morphological variability is frequently constrained, characteristics may not express at all the phases of plant development and appearance may be influenced by external factors. The therapeutic potential of medicinal plants will be compromised by incorrect plant identification, affecting the human health. There have been numerous reports of toxic cases, most of which were caused by species misidentification (Viljoen 2013). The issue of an increase in herbal treatments that have been tampered with or replaced with other plant materials has highlighted the necessity of quality monitoring (Raterta et al. 2014).
With regard to DNA level identification, molecular technologies provide much more useful informations. Nowadays, several PCR based approaches could be used to distinguish between species of the same genus. Today, genetic marker technology has been used by many researchers for studying genetic resources as the conventional methods have their limitations. A perfect DNA barcode should have enough conserved segments to design a universal primer, high variability to be used for species discrimination and the ability to differentiate among the closely related species. This is possible if a species has a very large genetic distance from the other intraspecific members of the group (Hebert et al. 2004; Mankga et al. 2013 , Cahyaningsih et al 2022).
DNA barcode research has been extensively used for phylogenetic tree-based methods to assign species to their appropriate taxa. The most popular phylogenetic tree is NJ, and the main criteria for evaluation were morphological distance, evolutionary history and species documentation (Liu et al. 2014). Today researchers working in the field of plant sciences have access to a variety of DNA markers. However, the difficulty of developing a universal barcode for the identification of all plant species has been contested by several researchers, and it is because of morphological and geographical variations as well as reticulate evolution (Mosa et al. 2019).
According to a review on the role of DNA barcoding as a powerful tool for plant biodiversity analysis, the ITS and rbcL genes have been acknowledged as fundamental barcode markers (Mosa et al. 2019). This further supports our assessment of ‘ITS’ dependability as a DNA barcode marker. However, there is no standardised barcode for plant identification, therefore researchers are using different sets of marker genes to increase the accuracy of species identification, (Cahyaningsih et al 2022).
CONCLUSION
The Polygonum posumbu collected from Awang- khunou, Manipur, India was successfully identified based on DNA characterization at ITS gene. The current study suggests that ITS is the reliable marker for identification of Polygonum posumbu from Manipur, India using the BLAST analysis genetic distance approach as deduced by phylogenetic tree. Our study for the identification of Polygonum posumbu plant from this region will help in further research on the important properties of this particular plant.
Ethics approval and consent to participate: Not applicable
Conflict of interests: Authors declare no conflict of interests to disclose
ACKNOWLEDGEMENTS
The authors gratefully acknowledge the financial support of IBSD, Imphal, and Department of Biotechnology (DBT), Govt. of India. The authors are also thankful to lab members for their continuous support.
REFERENCES
Alvarez I, Wendel JF (2003). Ribosomal ITS sequences and plant phylogenetic inference. Molecular Phylogenetics and Evolution, 29, 417– 434, doi: 10.1016/s1055-7903(03)00208-2.
Asha Devi, S Manjula, KR Shankaranarayana Rao, BS (2011). Healthy brain and well-being: sedentary lifestyle can impact cognitive ability adversely. In Bergin, M.G., (Ed.), Sedentary behavior: physiology, health risk, and interventions (pp. 1-27). Nova Inc.
Baldwin BG, Sanderson MJ, Porter JM, et al. (1995). The ITS region of nuclear ribosomal DNA – a valuable source of evidence on angiosperm phylogeny. Annals of the Missouri Botanical Garden, 82, 247–277, https://doi.org/10.2307/2399880
Buchheim MA, Keller A, Koetschan C et al. (2011). Internal transcribed spacer 2 (nu ITS2 rRNA) sequence-structure phylogenetics: towards an automated reconstruction of the green algal tree of life. PLoS ONE, 6, e16931, doi: 10.1371/journal.pone.0016931.
Cahyaningsih R Lindsey Jane Compton, Sri Rahayu, Joana Magos Brehmand Nigel Maxted (2022) DNA Barcoding Medicinal Plant Species from Indonesia Plants 2022, 11(10), 1375; https://doi.org/10.3390/plants11101375
CBOL, (2009). Plant working group a DNA barcode for land plants. Proc. Natl. Acad. Sci. USA106, 12794–12797, doi: 10.1073/pnas.0905845106
Chase MW, Fay MF (2009). Barcoding of plants and fungi. Science, 325, 682–683, DOI: 10.1126/SCIENCE.1176906
Fazekas AJ, Kesanakurti PR, Burgess KS et al. (2009). Are plant species inherently harder to discriminate than animal species using DNA barcoding markers ? Molecular Ecology Resources, 9, 130–139, DOI: 10.1111/J.1755-0998.2009.02652.X
Gao T, Pang XH, Chen SL (2009). Authentication of plants in Astragalus by DNAbarcoding technique. Planta Med. http://dx.doi.org/10.1055/s-0029-1234433
Hebert PDN, Penton EH, Burns JM, Janzen DH, Hallwachs W (2004). Tenspecies in one: DNA barcoding reveals cryptic species in the Neotropical skipperbutterfly Astraptes fulgerator. Proc. Natl. Acad. Sci. U.S.A. 101, 14812–14817, DOI: 10.1073/PNAS.0406166101
Hillis DM, Dixon MT, (1991). Ribosomal DNA – molecular evolution and phylogenetic inference. Quarterly Review of Biology, 66, 411–453 doi: 10.1086/417338.
Hollingsworth PM, Graham SW, Little DP (2011). Choosing and using a plant DNA barcode. PLoS One;6:e19254 , doi: 10.1371/journal.pone.0019254.
Ishwori L, Anupam DT, Singh PK, Dutta CM, Deepa N (2014). Antibacterial activity of some selected plants traditionally used as medicine in Manipur. African Journal of Biotechnology, 13, 1491-1495, doi: 10.5897/ajb2013.13495
Keller A, Wolf M, Dandekar T (2010). Ribosomal RNA phylogenetics: the third dimension. Biologia, 65, 388–391, https://doi.org/10.2478/s11756-010-0045-3
Kress WJ, Wurdack KJ, Zimmer EA, Weigt LA, Janzen DH (2005) . Use of DNA barcodes to identify flowering plants. Proc Natl AcadSci U S A;102:8369-74, doi: 10.1073/pnas.0503123102
Leishangthem S, Dinendra SL (2014). Study of some medicinal plants found in Imphal-East, District, Manipur, India. International Journal of Science and Research, Publications, 4. ISSN 2250-3153.
Li DZ, Gao LM, Li HT et al. (2011). Comparative analysis of a large dataset indicates that internal transcribed spacer (ITS) should be incorporated into the core barcode for seed plants. Proceedings of the National Academy of Sciences of the United States of America.108, 19641– 19646, DOI: 10.1073/PNAS.1104551108. EPUB 2011 NOV 18.
Li X, Wang B, Han R, Zheng Y, Yin HY, Xu L (2013). Identification of medicinal plants Chisandra chinensis using a potential DNA barcode ITS2. Acta Soc. Bot. Pol. 82,283–288.
Liu XF, Yang CH, Han HL, Ward RD, Zhang AB (2014). Identifying species of moths (Lepidoptera) from Baihua Mountain, Beijing, China, using DNAbarcodes. Ecol. Evol. 4, 2472–2487, https://doi.org/10.1002/ece3.1110
Liu Z, Chen SL, Song JY, Zhang SJ, Chen KL (2012). Applicationof deoxyribonucleic acid barcoding in Lauraceae plants. Pharmacogn Mag;8:4-11. doi: 10.4103/0973-1296.93301.
Mankga LT, Yessoufou K, Moteetee AM, Daru BH, Van der Bank M (2013). Efficacy of the core DNA barcodes in identifying processed and poorly conserved plant materials commonly used in South African traditional medicine. Zookeys 365, 215–233, DOI: 10.3897/ZOOKEYS.365.5730
Mosa KA, Gairola S, Jamdade R, El-Keblawy A, Al Shaer KI, Al Harthi EK, Shabana HA, Mahmoud T, (2019). The Promise of molecular and genomic techniques for biodiversity research and DNA barcoding of the Arabian Peninsula flora. Front. Plant Sci. 9 (1929), doi.org/10.3389/fpls.2018.01929.
Newmaster SG, Ragupathy S (2010). Ethnobotany genomics -discovery and innovation in a new era of exploratory research. J.Ethnobiol Ethnomed;6:2.
Pang X, Chen, S., 2014. Identification of medicinal plants using DNA barcoding technique. Encyc. Anal. Chem., 1–4, doi:10.1002/9780470027318.a9935.
Rao NK (2004). Plant genetic resources. Advancing conservation and use through biotechnology. Afr. J. Biotechnol. 3:136-145, doi: 10.5897/ajb2004.000-2025.
Reshmi Singha H, Kripamoy Chakraborty, Abhijit Datta (2016). An Overview of Medicinally Important Phyto Resources Used by the Manipuri Community of North Tripura District of Tripura, North East India. International Journal of Current Research in Biosciences and Plant Biology, 3(5), 46-53, doi: http://dx.doi.org/10.20546/ijcrbp.2016.305.007
Raterta R, Cabelin VLD, Alejandro GJD (2014). Molecular authentication of selected commercially sold medicinal plants in Quiapo, Manila. Philippines. Int.J. Sci. Tech. Res. 3, 22–26.
Rout GR, Mohapatra A (2008). Use of molecular markers in ornamental plants: A critical reappraisal. Eur. J. Hort. Sci. 37
Roy S, Tyagi A, Shukla V et al. (2010).Universal plant DNA barcode loci may not work in complex groups: a case study with Indian Berberis Species. PLoSONE 5, e13674, DOI: 10.1371/JOURNAL.PONE.0013674.
Schori M, Showalter AM (2011). DNA Barcoding as a means for identifying medicinal plants of Pakistan. Pak. J. Bot. 43, 1–4.
Siew YY, Zareisedehizadeh S, Seetoh WG, Neo SY, Tan CH, Koh HL (2014). Ethnobotanical survey of usage of fresh medicinal plants in Singapore. J Ethnopharm. 155, 1450–1466, doi: 10.1016/j.jep.2014.07.02.
Singh HB, Singh RS, Sandhu JS (2003). Herbal medicine of Manipur (A colorEncyclopedia). Daya publishing house, Delhi, (Volume 1).
Solmaz I, Sari N, Aka-Kacar Y and Yalcin-Mendi NY (2010). The genetic characterization of Turkish watermelon (Citrullus lanatus) accession using RAPD markers. Genetic Resources and Crop Evolution 57: 763–771, doi:10.1007/s10722-009-9515-2.
Song J, Shi L, Li D et al. (2012). Extensive pyrosequencing reveals frequent intra-genomic variations of internal transcribed spacer regions of nuclear ribosomal DNA. PLoS ONE, 7, e43971, DOI: 10.1371/JOURNAL.PONE.0043971.
Staggemeier VG, Diniz-Filho JA, Forest F, Lucas E (2015). Phylogenetic analysis in Myrcia section Aulomyrcia and inferences on plant diversity in the Atlantic rainforest. Annals of Botany, 115, 747–761, doi: 10.1093/aob/mcv005.
Stoeckle MY, Gamble CC, Kirpekar R, Young G, Ahmed S, Little DP (2011). Commercial teas highlight plant DNA barcode identification successes and obstacles. Sci Rep;1:42.
Viljoen AM (2013). Standardization and quality control of herbal medicinal products-does vibrational spectroscopy offer the solution? J. Med. Plant. Nat.Prod. Res. 79, 369–421, doi:10.1055/s-0033-1336434.
Yuan QJ, Zhang B, Jiang D et al. (2015). Identification of species and materia medica within Angelica L. (Umbelliferae) based on phylogeny inferred from DNA barcodes. Molecular Ecology Resources, 15, 358– 371, doi: 10.1111/1755-0998.12296.
Yumnam JY, Tripath OP(2012). Traditional knowledge of eating raw plants by the meitei of Manipur as medicine/nutrient supplement in their diet. Indian journal of traditional knowledge, 11, 1.
Zhou J, Wang W, Liu M, Liu Z (2014). Molecular authentication of the traditional medicinal plant Peucedanumpra eruptorum and its substitutes and adulterants by DNA -barcoding technique. Pharmacogn. Mag. 10, 385–390, doi: 10.4103/0973-1296.141754.


