Animal Cytogenetics Lab, Department of Zoology, University of Jammu, India
Corresponding author email: ramanjasrotia18@gmail.com
Article Publishing History
Received: 07/12/2020
Accepted After Revision: 25/03/2021
The Union Territory of Jammu & Kashmir have rich faunal diversity in its aquatic resources. Shellfishes (such as prawns and crabs) together with finfishes are contributing significantly to meet the nutritional requirements of natives. The local prawns have greater potential to raise the economic standard of Jammu region if cultured extensively on commercial scale. In this regard, they need to be analysed at chromosomal and molecular level. In the present study, the chromosomes of Himalayan prawn (Macrobrachium dayanum) were characterized by means of conventional Giemsa staining, Ag-NOR and G-banding techniques. It is one of the most abundant shellfishes in water bodies of Jammu region having high protein and mineral content. The diploid chromosome number (2n) and fundamental number (NF) were found to be 100 and 176 respectively. The karyotype comprised of 60 metacentric, 16 submetacentric, 12 subtelocentric and 12 telocentric chromosomes. Idiograms were constructed on the basis of morphometric details of the chromosomes. Allosomes (sex chromosomes) remained indistinguishable. NORs were located on two submetacentric pairs of the complement. Results of G-banding provided the heterochromatin and euchromatin patterns of M. dayanum. Several meiotic stages such as leptotene, zygotene, pachytene, diplotene, diakinesis, metaphase I and metaphase II from testes were also observed. Karyological studies aid in exact taxonomic identification and understanding of the phylogeny of an organism. The data obtained in present work would serve the basis of stock improvement, future cross breeding and chromosomal manipulation experiments such as induction of polyploidy etc. Through this analysis we have concluded the results which can support future research by acting as a credible milestone.
Chromosomes, G-banding, Ag-NOR, karyotype, M. dayanum, Metaphase
Jasrotia R, Langer S. Karyotypic Analysis and Chromosome Banding in Freshwater Prawn Macrobrachium dayanum from Jammu and Kashmir. Biosc.Biotech.Res.Comm. 2021;14(1).
Jasrotia R, Langer S. Karyotypic Analysis and Chromosome Banding in Freshwater Prawn Macrobrachium dayanum from Jammu and Kashmir. Biosc.Biotech.Res.Comm. 2021;14(1). Available from: <a href=”https://bit.ly/3au0rNh”>https://bit.ly/3au0rNh</a>
Copyright © This is an Open Access Article distributed under the Terms of the Creative Commons Attribution License (CC-BY). https://creativecommons.org/licenses/by/4.0/, which permits unrestricted use distribution and reproduction in any medium, provided the original author and sources are credited.
INTRODUCTION
Macrobrachium dayanum belongs to family Palaemonidae of decapod crustaceans. It is broadly distributed in Northern India, Southern Nepal and Myanmar (Jayachandran, 2001; Cai and Ng, 2002). It is commonly available prawn in stream ecosystems of Jammu region and its nutritional value stands at par with culturable fish species. The identifying features of the species are: Rostrum straight or slightly upturned at distal half, reaching almost equal to the length of antennal scale or extending a little beyond it. Dorsal or upper surface of the rostrum bears 5-11 teeth of which 1-2 are post orbital and the ventral or lower surface possess 4-7 teeth (Paul, 1991; Sharma, 2015). Sexual dimorphism is quite distinct in M. dayanum. Second pair of walking leg is stout and more robust with sharp pincers in males as compared to females. The second pair of swimmerets bears an additional structure called appendix masculina in males. The size of the specimen ranged from 5.0±0.10 to 6.3±0.38 cm in males and 4.9±0.12 to 5.9±0.36 cm in females. The females carry green coloured eggs in the brood chamber during the breeding season (Langer et al., 2004; Jasrotia et al., 2017; Jasrotia and Langer, 2019).
Despite having greater economic and commercial importance, the cytogenetic reports of family Palaemonidae in general and genus Macrobrachium in particular are very few. The reason for this is attributed to the technical difficulties associated with their highly condensed numerous chromosomes (Chow et al., 1990; Nagashree, 1993; Gonzalez-Tizon et al., 2013; Phimphan et al., 2018). There is no previous record of the karyotype of M. dayanum. The present study was thus undertaken to document the chromosome number, analysis of meiotic stages, development of karyotype and chromosomal banding of this species for the first time. It is pertinent to mention that karyomorphological information contributes to better understanding of systematics and genealogy. Moreover, it would help in analysing the course of evolution in family Palaemonidae (Phimphan et al., 2018).
MATERIAL AND METHODS
Live specimens of M. dayanum were collected by using cast net from Gho-manhasan stream and Sai stream of Jammu district and brought to Animal Cytogenetics lab, Department of Zoology, University of Jammu in the plastic containers. The taxonomic identification of specimens is based on the standard keys. Before dissection, the animals were maintained in clean water in glass troughs equipped with aerators and thermoregulators (Jayachandran, 2001; Cai and Ng, 2002; Sharma, 2015). Adult specimens were injected intramuscularly with 0.05 % colchicine solution and were maintained for a period of 5 hours before sacrifice. Apart from this, dip treatment of 0.1 % colchicine solution for 10-12 hours was also applied on some specimens. Gonadal tissues, hepatopancreas and fertilized eggs were used for chromosomal preparations by following air-drying Giemsa staining technique with some modifications (Choudhary et al., 2013; Hassan et al., 2015). After colchicine treatment, the prawns were dissected and the required tissues were placed in hypotonic solution (0.9 % sodium citrate) for 50 minutes (Sharma, 2015).
Fixation of the tissue was done in 3:1 methanol-acetic acid fixative (Carnoy’s fixative) for 60 minutes (with three changes of fixative after every 20 minutes). The material was then minced in 45% acetic acid for 10-15 minutes. The suspension was dropped on the clean and pre-warmed slides and air dried. The conventional method of dabbing the fixed tissue material on clean slides followed by air drying was also used. After air-drying, the slides were stained with 4% Giemsa phosphate buffer solution (pH 6.8) for 30- 35 minutes. Ag-NOR and G-banding were done following standard protocols with certain modifications (Howell and Black, 1980; Sumner et al., 1971). The prepared slides were scanned under Olympus camera aided microscope and metaphase spreads as well as meiotic stages were photographed using Sony SSC-DC378P camera under 1000x magnification. For karyotyping, best metaphase spreads were selected and chromosomes were classified following internationally accepted standard classification (Levan et al., 1964). The chromosomal pairs were arranged in the decreasing order of their size in the karyogram. Morphometric measurements were done by using occulometer (Hassan et al., 2015).
RESULTS AND DISCUSSION
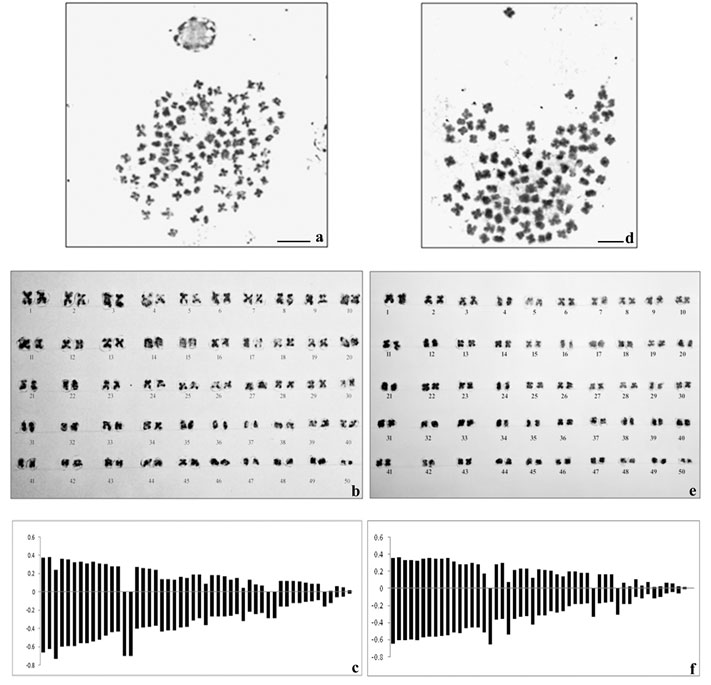
The spermatagonial metaphase (Fig.1a) in male and somatic metaphase complement in female (Fig.1d) comprised of 50 chromosome pairs in each showing basic chromosome number to be 2n=100 in this species. The chromosome type and form were found to be similar in both the sexes and most of the chromosomes were metacentric and sub-metacentric. The diploid chromosome formula was determined as 2n=60m+16sm+12st+12t. Sex chromosomes were not morphologically differentiated from the autosomes in male and female karyotypes (Fig. 1b and 1e respectively). The average lengths of each chromosome including short and long arm length, total length, arm ratio, relative length percentage and centromeric index were calculated for both the sexes and presented in Table 1 and 2. The diagrammatic summary of male and female karyotype was shown by constructing the idiograms (Fig.1c and 1f).
Figure 1: Metaphase complements (2n=100), karyotypes and idiograms of Macrobrachium dayanum a. Spermatogonial metaphase (male) b. Karyotype of male c. Idiogram of male d. Metaphase plate (female) e. Karyotype of female f. Idiogram of female, Bars=5 µm.
Table 1. Karyomorphometric data of Macrobrachium dayanum (male)
| Chromosome pair No. | Mean length of Short arm (p) (µm) | Mean length of Long arm (q) (µm) | Absolute length (p+q) (µm) | Arm ratio (q/p) | Relative length %age | Centromeric index | Nomenclature |
| 1. | 0.37 | 0.66 | 1.03 | 1.78 | 3.98 | 35.9 | Sub-metacentric |
| 2. | 0.38 | 0.62 | 1.0 | 1.63 | 3.87 | 38 | Metacentric |
| 3. | 0.24 | 0.73 | 0.97 | 3.04 | 3.75 | 24.7 | Sub-telocentric |
| 4. | 0.36 | 0.60 | 0.96 | 1.66 | 3.71 | 37.5 | Metacentric |
| 5. | 0.35 | 0.59 | 0.94 | 1.68 | 3.64 | 37.2 | Metacentric |
| 6. | 0.32 | 0.59 | 0.91 | 1.84 | 3.52 | 35.16 | Sub-metacentric |
| 7. | 0.33 | 0.56 | 0.89 | 1.69 | 3.44 | 37.07 | Metacentric |
| 8. | 0.31 | 0.56 | 0.87 | 1.80 | 3.36 | 35.63 | Sub-metacentric |
| 9. | 0.33 | 0.54 | 0.87 | 1.63 | 3.36 | 37.9 | Metacentric |
| 10. | 0.31 | 0.52 | 0.83 | 1.67 | 3.21 | 37.3 | Metacentric |
| 11. | 0.30 | 0.48 | 0.78 | 1.6 | 3.02 | 38.4 | Metacentric |
| 12. | 0.28 | 0.44 | 0.72 | 1.57 | 2.78 | 38.8 | Metacentric |
| 13. | 0.28 | 0.43 | 0.71 | 1.53 | 2.74 | 39.4 | Metacentric |
| 14. | – | 0.70 | 0.70 | – | 2.71 | – | Telocentric |
| 15. | – | 0.70 | 0.70 | – | 2.71 | – | Telocentric |
| 16. | 0.27 | 0.40 | 0.67 | 1.48 | 2.59 | 40.29 | Metacentric |
| 17. | 0.26 | 0.39 | 0.65 | 1.5 | 2.51 | 40 | Metacentric |
| 18. | 0.25 | 0.38 | 0.63 | 1.52 | 2.43 | 39.6 | Metacentric |
| 19. | 0.24 | 0.37 | 0.61 | 1.54 | 2.36 | 39.3 | Metacentric |
| 20. | 0.14 | 0.43 | 0.57 | 3.07 | 2.20 | 24.5 | Sub-telocentric |
| 21. | 0.14 | 0.42 | 0.56 | 3.0 | 2.16 | 25 | Sub-metacentric |
| 22. | 0.13 | 0.42 | 0.55 | 3.23 | 2.13 | 23.6 | Sub-telocentric |
| 23. | 0.16 | 0.39 | 0.55 | 2.43 | 2.13 | 29.09 | Sub-metacentric |
| 24. | 0.15 | 0.38 | 0.53 | 2.53 | 2.05 | 28.3 | Sub-metacentric |
| 25. | 0.19 | 0.31 | 0.50 | 1.63 | 1.93 | 38 | Metacentric |
| 26. | 0.19 | 0.29 | 0.48 | 1.52 | 1.85 | 39.5 | Metacentric |
| 27. | 0.09 | 0.36 | 0.45 | 4 | 1.74 | 20 | Sub-telocentric |
| 28. | 0.18 | 0.27 | 0.45 | 1.5 | 1.74 | 40 | Metacentric |
| 29. | 0.18 | 0.27 | 0.45 | 1.5 | 1.74 | 40 | Metacentric |
| 30. | 0.17 | 0.27 | 0.44 | 1.58 | 1.70 | 38.6 | Metacentric |
| 31. | 0.13 | 0.26 | 0.39 | 2.0 | 1.51 | 33.3 | Sub-metacentric |
| 32. | 0.15 | 0.24 | 0.39 | 1.6 | 1.51 | 38.4 | Metacentric |
| 33. | 0.04 | 0.32 | 0.36 | 8 | 1.39 | 11.12 | Telocentric |
| 34. | 0.13 | 0.22 | 0.35 | 1.69 | 1.35 | 37.14 | Metacentric |
| 35. | 0.08 | 0.24 | 0.32 | 3 | 1.23 | 25 | Sub-metacentric |
| 36. | 0.07 | 0.23 | 0.30 | 3.2 | 1.16 | 23.3 | Sub-telocentric |
| 37. | – | 0.29 | 0.29 | – | 1.12 | – | Telocentric |
| 38. | – | 0.29 | 0.29 | – | 1.12 | – | Telocentric |
| 39. | 0.12 | 0.16 | 0.28 | 1.33 | 1.08 | 42.8 | Metacentric |
| 40. | 0.12 | 0.16 | 0.28 | 1.33 | 1.08 | 42.8 | Metacentric |
| 41. | 0.12 | 0.12 | 0.24 | 1 | 0.92 | 50 | Metacentric |
| 42. | 0.11 | 0.12 | 0.23 | 1.09 | 0.89 | 47.8 | Metacentric |
| 43. | 0.10 | 0.11 | 0.21 | 1.1 | 0.81 | 47.6 | Metacentric |
| 44. | 0.09 | 0.10 | 0.19 | 1.1 | 0.73 | 47.3 | Metacentric |
| 45. | 0.09 | 0.09 | 0.18 | 1 | 0.69 | 50 | Metacentric |
| 46. | – | 0.16 | 0.16 | – | 0.61 | – | Telocentric |
| 47. | 0.01 | 0.12 | 0.13 | 6 | 0.50 | 7.69 | Sub-telocentric |
| 48. | 0.06 | 0.06 | 0.12 | 1 | 0.46 | 50 | Metacentric |
| 49. | 0.05 | 0.05 | 0.10 | 1 | 0.38 | 50 | Metacentric |
| 50. | 0.02 | 0.02 | 0.04 | 1 | 0.15 | 50 | Metacentric |
Morphometric measurement of the chromosomes showed mean haploid length to be 25.82 µm and 25.79 µm in male and female respectively. The total complement length was recorded as 51.64 µm in male and 51.58 µm in female. The absolute length of the largest chromosome was 1.03 µm and that of the smallest chromosome was 0.04 µm in male whereas absolute length of the largest chromosome was 1.0 µm and that of the smallest chromosome was 0.01 µm in female. Centromeric index for the largest and the smallest chromosome in male was calculated as 35.9 and 50 respectively. However, the CI for the largest and the smallest chromosome in female was found to be 35and 50 respectively.
Table 1. Karyomorphometric data of Macrobrachium dayanum (female)
| Chromosome pair No. | Mean length of Short arm (p) (µm) | Mean length of Long arm (q) (µm) | Absolute length (p+q) (µm) | Arm ratio (q/p) | Relative length %age | Centromeric index | Nomenclature |
| 1. | 0.35 | 0.65 | 1.0 | 1.85 | 3.87 | 35 | Sub-metacentric |
| 2. | 0.36 | 0.61 | 0.97 | 1.69 | 3.76 | 37.1 | Metacentric |
| 3. | 0.33 | 0.61 | 0.94 | 1.84 | 3.64 | 35.1 | Sub-metacentric |
| 4. | 0.33 | 0.60 | 0.93 | 1.81 | 3.60 | 35.4 | Sub-metacentric |
| 5. | 0.32 | 0.61 | 0.93 | 1.90 | 3.60 | 34.4 | Sub-metacentric |
| 6. | 0.34 | 0.58 | 0.92 | 1.70 | 3.56 | 36.9 | Metacentric |
| 7. | 0.35 | 0.57 | 0.92 | 1.62 | 3.56 | 38.04 | Metacentric |
| 8. | 0.34 | 0.57 | 0.91 | 1.67 | 3.52 | 37.3 | Metacentric |
| 9. | 0.34 | 0.56 | 0.90 | 1.64 | 3.48 | 37.7 | Metacentric |
| 10. | 0.35 | 0.55 | 0.90 | 1.57 | 3.48 | 38.8 | Metacentric |
| 11. | 0.31 | 0.52 | 0.83 | 1.67 | 3.21 | 37.3 | Metacentric |
| 12. | 0.28 | 0.53 | 0.81 | 1.89 | 3.14 | 34.5 | Sub-metacentric |
| 13. | 0.28 | 0.47 | 0.75 | 1.67 | 2.90 | 37.3 | Metacentric |
| 14. | 0.29 | 0.46 | 0.75 | 1.58 | 2.90 | 38.6 | Metacentric |
| 15. | 0.28 | 0.46 | 0.74 | 1.64 | 2.86 | 37.8 | Metacentric |
| 16. | 0.17 | 0.52 | 0.69 | 3.05 | 2.67 | 24.6 | Sub-telocentric |
| 17. | – | 0.66 | 0.66 | – | 2.55 | – | Telocentric |
| 18. | 0.28 | 0.37 | 0.65 | 1.32 | 2.52 | 43.07 | Metacentric |
| 19. | 0.29 | 0.36 | 0.65 | 1.24 | 2.52 | 44.61 | Metacentric |
| 20. | 0.07 | 0.54 | 0.61 | 7.71 | 2.36 | 11.4 | Telocentric |
| 21. | 0.21 | 0.36 | 0.57 | 1.714 | 2.21 | 36.8 | Sub-metacentric |
| 22. | 0.23 | 0.34 | 0.57 | 1.47 | 2.21 | 40.3 | Metacentric |
| 23. | 0.23 | 0.33 | 0.56 | 1.43 | 2.17 | 41.07 | Metacentric |
| 24. | 0.12 | 0.43 | 0.55 | 3.58 | 2.13 | 21.8 | Sub-telocentric |
| 25. | 0.22 | 0.32 | 0.54 | 1.45 | 2.09 | 40.7 | Metacentric |
| 26. | 0.21 | 0.32 | 0.53 | 1.52 | 2.05 | 39.6 | Metacentric |
| 27. | 0.20 | 0.27 | 0.47 | 1.35 | 1.82 | 42.5 | Metacentric |
| 28. | 0.16 | 0.29 | 0.45 | 1.81 | 1.74 | 35.5 | Sub-metacentric |
| 29. | 0.14 | 0.27 | 0.41 | 1.92 | 1.58 | 34.1 | Sub-metacentric |
| 30. | 0.19 | 0.21 | 0.40 | 1.10 | 1.55 | 47.5 | Metacentric |
| 31. | 0.19 | 0.19 | 0.38 | 1 | 1.47 | 50 | Metacentric |
| 32. | 0.18 | 0.19 | 0.37 | 1.05 | 1.43 | 48.6 | Metacentric |
| 33. | 0.18 | 0.18 | 0.36 | 1 | 1.39 | 50 | Metacentric |
| 34. | – | 0.34 | 0.34 | – | 1.31 | – | Telocentric |
| 35. | 0.16 | 0.18 | 0.34 | 1.12 | 1.31 | 47 | Metacentric |
| 36. | 0.16 | 0.17 | 0.33 | 1.06 | 1.27 | 48.4 | Metacentric |
| 37. | 0.16 | 0.16 | 0.32 | 1 | 1.24 | 50 | Metacentric |
| 38. | – | 0.31 | 0.31 | – | 1.20 | – | Telocentric |
| 39. | 0.06 | 0.19 | 0.25 | 3.16 | 0.96 | 24 | Sub-telocentric |
| 40. | 0.02 | 0.19 | 0.21 | 9.5 | 0.81 | 9.52 | Telocentric |
| 41. | 0.10 | 0.10 | 0.20 | 1 | 0.77 | 50 | Metacentric |
| 42. | 0.03 | 0.12 | 0.15 | 4 | 0.58 | 20 | Sub-telocentric |
| 43. | 0.07 | 0.07 | 0.14 | 1 | 0.54 | 50 | Metacentric |
| 44. | 0.01 | 0.12 | 0.13 | 12 | 0.50 | 7.69 | Telocentric |
| 45. | 0.02 | 0.10 | 0.12 | 5 | 0.46 | 16.6 | Sub-telocentric |
| 46. | 0.06 | 0.06 | 0.12 | 1 | 0.46 | 50 | Metacentric |
| 47. | 0.05 | 0.05 | 0.10 | 1 | 0.38 | 50 | Metacentric |
| 48. | 0.019 | 0.06 | 0.079 | 3.15 | 0.30 | 24 | Sub-telocentric |
| 49. | 0.015 | 0.015 | 0.03 | 1 | 0.11 | 50 | Metacentric |
| 50. | 0.005 | 0.005 | 0.01 | 1 | 0.03 | 50 | Metacentric |
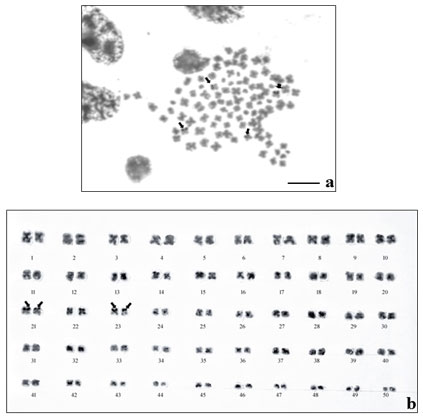
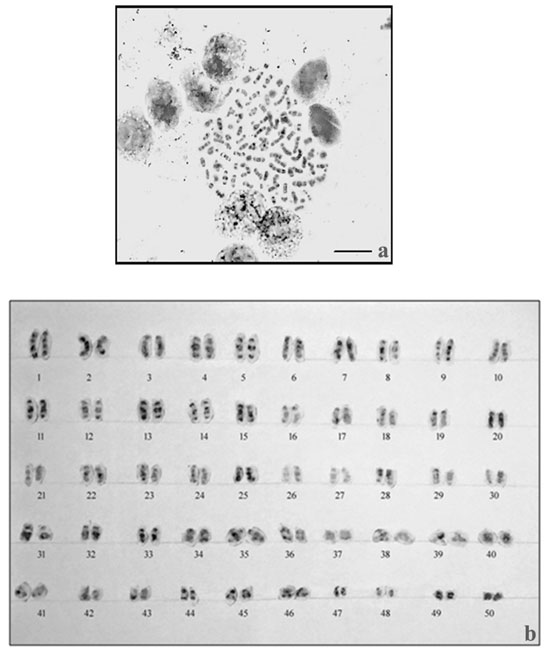
The results of NOR- banding revealed the presence of NORs on two submetacentric pairs of NOR banded complement (Fig. 2a). NORs are associated with gene expressions. The NOR-banded karyotype is represented in figure 2b. By G-banding, a series of light and dark bands were produced that allow for the positive identification of each chromosome in the complement (Fig. 3a). The dark bands are A–T rich, heterochromatic regions of the chromosomes, while the light bands are C–G rich, euchromatic regions. The G-banded karyotype is respresented in figure 3b.
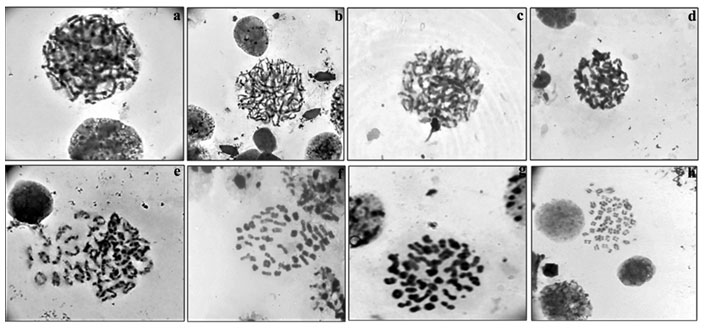
Among meiotic stages (Fig.4a-h) from testes, leptotene (characterized by network of chromosomes), zygotene (chromosomes with free ends and synapsis of homologous chromosomes was observed), pachytene (chromosomes were slightly more condensed than in zygotene), diplotene (chromosomes with morphology of number eight and plus shaped indicating the places of cross over exchanges), diakinesis (chromosomes were further condensed and have assumed morphology of rings marking the chiasmata terminalisation) and metaphase II (with 50 chromosomes) were clearly visible.
Figure 2: NOR-banding in M. dayanum a. NOR-banded metaphase complement (Arrows indicating the NOR regions) b.NOR-banded karyotype of M. dayanum (NOR bands on two sub-metacentric pairs)
The chromosomes of prawns of family Palaemonidae are not only very small in size and large in number but also showed a wide range of variations from species to species. The diploid number ranges from 56 in Palaemon serratus to 124 in Macrobrachium villosimanus (Chaudhary et al., 2013; Gonzalez-Tizon et al., 2013). However, except for M. carcinus (2n = 94), most Macrobrachium species possessed a diploid number either equal to or higher than 100 as Macrobrachium siwalikensis (2n = 100), M. nipponense (2n = 104), M. idella (2n = 104) and M. scabriculum (2n = 104), Palaemon lamarrei (2n = 118), M. rosenbergii (2n = 118), Macrobrachium villosimanus (2n = 124) (Mittal and Dhall, 1971; Vishnoi, 1972; Damrongphol et al., 1991; Qiu et al., 1994; Lakra and Kumar, 1995; Indy et al., 2009; Choudhary et al., 2013). The diploid number 2n=100 found in M. dayanum is consistent with the diploid number found in other congeneric species. NOR- and G-banding results of present study are found to be in accordance with the chromosomal banding analysis in Macrobrachium villosimanus and Macrobrachium lanchesteri (Choudhary et al., 2013; Phimphan et al., 2018).
Figure 3: G-banding in M. dayanuma. G-banded metaphase complement b. G-banded karyotype
Figure 4: Meiotic stages observed in testicular tissue of M. dayanum a. Leptotene b. Zygotene c. Early Pachytene d. Late Pachytene e. Diplotene f. Diakinesis g. MetaphaseI h. MetaphaseII
CONCLUSION
The present study is the first report on karyotype and chromosomal banding in Macrobrachium dayanum from UT of J&K. The diploid number observed to be 100 with the karyotypic formula 60m+16sm+12st+12t. Numerous gene expressions regions i.e. NORs were located on two submetacentric pairs by silver staining. Alternate light and dark bands on chromosomes depicting GC and AT rich regions were revealed by G banding. The present work will serve as baseline for the genetic improvement, hybridisation experiments, conservation and management programmes for Macrobrachium dayanum. The data obtained in current study will help the researchers in prawn systematics for the valid species identification. Further fluorescence in situ hybridisation and molecular studies like mitochondrial DNA analysis, 16S rRNA analysis, microsatellite analysis and DNA sequencing would strengthen the field of prawn genetics in Jammu region.
ACKNOWLEDGEMENTS
The authors would like to thank Head, Department of Zoology, University of Jammu, Jammu India for providing necessary facilities to conduct the experiment.
Conflict of Interest: The authors declare no conflict of interest.
Conflict of Interest: The authors declare no conflict of interest.
Ethical Clearance Statement: The Current Research Work Was Ethically Approved by the Institutional Review Board (IRB) of University of Jammu, India.
REFERENCES
Cai, Y. and Ng, P.K.L. (2002). The freshwater palaemonid prawns (Crustacea: Decapoda: Caridea) of Myanmar. Hydrobiologia, 487: 59-83.
Choudhary, N., Sharma, R., Asthana, S., Vyas, P., Rather, M.A., Reddy, A.K. and Krishna, G. (2013). Development of Karyotype and Localisation of Cytogenetic Markers in Dimua River Prawn, Macrobrachium villosimanus (Tiwari, 1949). Journal of Biological Sciences, 13(6): 507-513.
Chow, S., Dougherty, W.J. and Sandifer, P.A. (1990). Meiotic chromosome complements and nuclear DNA contents of four species of shrimps of the genus Penaeus. Journal of Crustacean Biology. 10(1): 29–36.
Damrongphol, P., Eangchuan, N., Ajpru, S., Poolsanguan, B. and Withyachumnarnkul, B. (1991). Karyotype of the giant freshwater prawn, Macrobrachium rosenbergii. Journal of the Science Society of Thailand. 17: 57-69.
González-Tizón, A.M., Rojo, V., Menini, E., Torrecilla, Z. and Martínez-Lage, A. (2013). Karyological analysis of the shrimp Palaemon serratus (Decapoda: Palaemonidae). Journal of Crustacean Biology, 33(6): 843-848.
Hassan, H., Leitao, A., Al-Shaikh, I. and Al-MaslaMani, I. (2015). Karyotype of Palaemon khori (Decapoda: Palaemonidae). Vie et milieu- Life and Environment, 65 (3): 151-155.
Howell, W.M. and Black, D.A. (1980). Controlled silver-staining of nucleolus organizer regions with a protective colloidal developer: a 1-step method. Experientia,36:1014–5.
Indy, J.R., Arias-Rodriguez, L., Paramo-Delgadillo, S., Hernandez-Guzman, J., D’ Artola Barcelo, A.L. and Contreras-Sanchez, W. (2009). Cytogenetic studies of invertebrate species from Tabasco, Mexico. Proceedings of the world Aquaculture Meeting, VeraCruz, Mexico.
Jasrotia, R. and Langer, S. (2019). Comparative Account of DNA Extraction Protocols in Some Freshwater Prawns of Genus Macrobrachium (Bate, 1868) (Family Palaemonidae) from Jammu Waters for PCR Based Applications. Biomedical and Pharmacology Journal 12(3): 1201-1206
Jasrotia, R., Langer, S., Palaq., Sharma, N. and Panjaliya, R.K. (2017). Assessment of genetic variability of two freshwater prawns Macrobrachium dayanum and Macrobrachium lamarrei from Jammu region by using ISSR markers. International Journal of Recent Scientific Research, 8(10): 21176-21180.
Jayachandran, K.V. (2001). Palaemonid prawns: Taxonomy, Biodiversity, Biology and Management. Science publishers, Inc., USA, 49-181. ew
Lakra, W.S. and Kumar, P. (1995). Studies on the chromosomes of the freshwater prawns Macrobrachium idella and M. scrabiculum (Crustacea Decapoda Palaemonidae). Cytobios, 84: 147-156.
Langer, S., Kour, T. and Bakhtiyar, Y. (2004). Studies on the effect of varying levels of dietary protein on growth and survival of freshwater prawn Macrobrachium dayanum. J. Aqua Biol., 19(1): 187-191.
Levan, A., Fredga, K. and Sandberg, A.A. (1964). Nomenclature for centromere position in chromosomes. Hereditas,52: 201-220.
Mittal, O.P. and Dhall, U. (1971). Chromosome Studies in Three Species of Freshwater Decapods (Crustacea). Cytologia, 36: 633-638.
Nagashree, N.S. (1993). Comparative studies on the chromosome complements of freshwater decapod crustaceans. Ph.D. Thesis, Bangalore University, Bangalore.
Paul, A.L. (1991). Distribution, ecology and biology of fresh water prawns (Macrobrachium spp.) of North Eastern region. Ph.D. Thesis, The North-Eastern Hill University, Shillong.
Phimphan, S., Tanomtong, A., Seangphan, N. and Sangpakdee, W. (2018). Chromosome studies on freshwater prawn, Macrobrachium lanchesteri (Decapoda, Palaemonidae) from Thailand. Nucleus. 62: 77-82.
Qiu, G., Du, N. and Lai, W. (1994). Chromosomal and karyological studies on the freshwater prawn Macrobrachium nipponense (Crustacea:Decapoda). Oceanol Limnol Sinica, 25: 493-498.
Sharma, N. (2015). Taxonomy and population dynamics of freshwater prawns inhabiting some Jammu waters. M.Phil. Dissertation, University of Jammu, Jammu.
Sumner, A.T., Evans, H.J. and Buckland, R.A. (1971). New technique for distinguishing between human chromosomes. Nature New Biology, 232: 31-32.
Vishnoi, D.N. (1972). Studies on the Chromosomes of Some Indian Crustacea. Cytologia, 37: 43-51.