Post Graduate and Research Department of Zoology, Chikkanna Government Arts
College, Tirupur, Tamil Nadu, India.
Corresponding author email: sarathkgm2010@gmail.com
Article Publishing History
Received: 26/09/2021
Accepted After Revision: 15/12/2021
Cholera is an acute infectious disease in countries with poor sanitation. The main clinical symptom of cholera is gastroenteritis and a symptom of the disease includes mild to moderate dehydration, vomiting, fever and abdominal pain. Antibiotic drug resistance in V. cholera has become a serious problem mostly in developing countries and muti-drug resistance to different antibiotics as well as Tetracycline, Ampicillin, Kanamycin, Trimethoprim, Sulphonamides Streptomycin and Gentamicin. Multi-drug resistant V. cholera showed resistance against three or more three contrasting classes of antibiotics for recent decades. Fifty diarrheal samples were collected from the different hospitals in and around the Tirupur district. 25 positive V. cholerae were isolated. The isolates were characterized morphologically and biochemically.
The confirmed strains were taken to decide susceptibility patterns by the disc diffusion method. Vibrio cholerae isolated in this study was subjected to an antibiogram against 10 commonly used antibiotics. All the tested isolates showed maximum resistance to Erythromycin (97%) and minimum resistance was noted in Trimethoprime (50%) respectively. Electrophoretic examination of plasmid DNA was carried out by Agarose gel Electrophoresis on 0.7%. More than 90% of resists showed were taken for plasmid isolation. The molecular weights of the fragments were evaluated by using a 10,000 bp DNA ladder and it was determined to be 1500 bp respectively. It is crucial to know the sensitivity and resistance pattern of any bacterial species for intercession an effective drug of choice in future.
Antibiotic Susceptibility Test, Diarrhea, Plasmid, Vibrio Cholerae.
kumar S, kumar A.M,. Isolation of Antibiotic Resistant Plasmid Deoxyribonucleic Acid from Diarrheal Pathogens. Biosc.Biotech.Res.Comm. 2021;14(4).
Kumar S, Kumar A.M,. Isolation of Antibiotic Resistant Plasmid Deoxyribonucleic Acid from Diarrheal Pathogens. Biosc.Biotech.Res.Comm. 2021;14(4). Available from: <a href=”https://bit.ly/3xX5IZc“>https://bit.ly/3xX5IZc</a>
Copyright © Kumar and Kumar This is an open access article distributed under the terms of the Creative Commons Attribution License (CC-BY) https://creativecommns.org/licenses/by/4.0/, which permits unrestricted use distribution and reproduction in any medium, provide the original author and source are credited.
INTRODUCTION
Cholera known as a global ‘disease of poverty’, is a bacterial gut infection producing liquefied diarrhea that can speedily lead to severe dehydration and prove fatal within hours, if untreated. Cholerae still continues to be the foremost cause of human infection, especially in developing countries that shortage of access to guarded drinking water and proper sanitation. It remains global intimidation and is one of the crucial pointers of social development. Presently World Health Organization (WHO) guesstimate, 1.3 – 4 million cholera cases globally every year and an estimated 21,000 – 143,000 deaths transpire (Sophia lonappan et al. 2020).
Nevertheless, the role of these islet in V. cholerae’s life cycle, predominantly the 26-kb VSP-II, remains badly understood (Murphy et al. 2021). The life-threatening diarrheal disease occurs in infestation form in many developing countries and it is one of the major public health loads with a high morbidity and mortality rate around the globe. Though important trouble has been made to identify the factual number of cholera cases, 90% – 95% of cases of cholera remain unheeded due to deficient surveillance systems and poor socio-economic conditions (Uddin et al. 2018; Anna et al. 2021).
Vibrio spp. infections generally do via the consumption of weakened seafood or exposure to polluted water and tend to be self-limiting (Anna et al. 2021). Over the years, a number of antimicrobials such as tetracycline, fluoroquinolones and azithromycin have been effectually used in the treatment of cholera patients (Das et al. 2020). The antibiotic resistance of V. cholerae strains has become a global warning problem, failing to treat the life of the patient. Recurrently MGEs play a central role in microbial evolution, ration as an apparatus by which hereditary material can be transferred, disseminated, and rearranged, allowing for rapid alteration to new and changing environments. At present, V. cholerae O1 was common among cases over five in a metro clinic in Bangladesh (Hamasalih et al. 2020).
Because its significant factors associated with antibiotic-resistant may be actionable in identifying patients with a high probability of multidrug-resistant (Stephanie et al. 2021). The soaring pathogenicity of Vibrio strains and general weakness of hospitalized patients make it necessary to assess the presence of strains in diarrheal samples. So hence the present research has made an attempt to point out the purpose of the susceptibility test were two folds; to guide the choice of the antibiotic treatment for the individual patients and provide scrutiny data to monitor resistance trends including the epidemiological data are to provide the resistant gene linked with plasmid.
MATERIAL AND METHODS
In this study, 50 diarrheal samples were collected from various hospitals in and around Tirupur District. The Isolation and identification of Vibrio species from the watery diarrhea sample were based on Thiosulfate Citrate Bile Salts Sucrose (TCBS) agar plates at 370C for 18-24 hours aerobically followed by staining and biochemical test which includes IMViC, Gram’s staining, Motility, Starch hydrolysis, Triple sugar iron, Ornithine Decarboxylase, Arginine Dihydrolase, Lysine, Decarboxylase and Gelatin test it is described by (Talukder et al. 2019).
Antibiotic Sensitivity Testing (AST) was performed by disc diffusion method using the antibiotics which include Ampicillin (10 mcg), Bacitracin (10 unit), Clindamycin (2 mcg), Erythromycin (15 mcg), Piperacillin (30 mcg), Ticarcillin (75 mcg) and Trimethoprim (5 mcg) respectively. The discs were gently placed into the MHA agar plates and the plates were incubated at 37o C for 24 hours. At the end of the incubation, the diameter of the inhibition zone was measured and calculated using a standard chart and detected the antibiotics as sensitive, intermediate, resistant to V. cholerae. Isolation of plasmids was performed by (Holmes and Quiley 1981). The electrophoresis probe of the plasmid DNA was carried out by Agarose Gel Electrophoresis on 0.7%. The molecular weight of the fragments was evaluated using a 1kb DNA ladder.
RESULTS AND DISCUSSION
Among the 50 diarrheal samples were collected from different Hospitals and Primary Health centers from rural areas of Kangayam around Tirupur city. The isolates were confirmed by using TCBS agar. Further, the identification of V. cholera isolates was done by a standard biochemical test (Fig I & II, Table I). The confirmed isolated were further taken to antimicrobial sensitivity test by disc diffusion method (Fig – III and Table -II). Erythromycin showed 97% and Trimethoprim showed minimum resistance (54%).
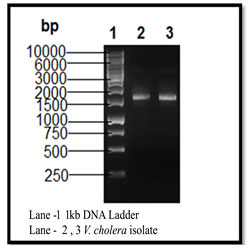
The maximum resistant pattern percentage (97%) was noted in strain no. NSKVC24 and the minimum resistance percentage (54%) were noted in strain-no NSKVC26 (Table – II). Electrophoretic probe of plasmid DNA was prepared and carried out by Agarose gel electrophoresis on 0.7%. Two isolates (NSKVC24 and NSKVC26) which showed more than 90% resistance against tested antibiotics were selected for plasmid isolation. The molecular size of the plasmid DNA was calculated to be 1500bp (Fig – IV) using a 1kb DNA ladder (Hamasalih et al. 2020).
Figure 1: V. cholerae on TCBS Agar
Figure 2: Biochemical Test

Figure 3: Antibiotic Susceptibility Test
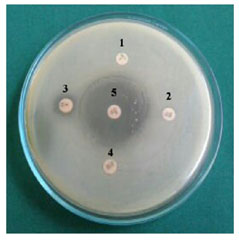
Table 1. Biochemical characterization of V. cholerae
| S. No. | Reaction | Result |
| 01. | Grams Staining | Negative |
| 02. | Indole | Positive |
| 03. | Methyl red | Negative |
| 04. | Voges proskauer | Positive |
| 05. | Citrate | Positive |
| 06. | Motility | Positive |
| 07. | Starch hydrolysis | Positive |
| 08. | Triple sugar iron agar | Negative |
| 09. | Ornithine Decarboxylase | Positive |
| 10. | Arginine Dihydrolase | Negative |
Table 2. A ntibiotic resistant percentage of V. cholerae
| S. No. | Name of the antibiotics | % of resistance |
| 01. | Ampicillin | 90 % |
| 02. | Bacitracin | 69 % |
| 03. | Clindamycin | 84 % |
| 04. | Erythromycin | 97 % |
| 05. | Piperacillin | 52 % |
| 06. | Ticarcillin | 90 % |
| 07. | Trimethoprime | 50 % |
Figure 4: Plasmid DNA isolation of V. cholerae
Globally, V. cholerae is recognized as an important bacterial pathogen contributing to hospital infection. Despite the use of potent antibiotics still, a high mortality rate exists in cases of cholera infection. In the present study, an antibiotic sensitivity test was done and the highest percentage of resistance was noted against the antibiotics tested, this is due to the development of antibiotic-resistant gene in the plasmid of V. cholerae. Resistance of V.
cholerae to antimicrobials is generally thought to be chromosomal in the inception and rarely plasmid-mediated because it is not very clear whether this pattern of plentiful resistance of Vibrio cholerae, to many antimicrobials, means that the organisms are obtaining more of plasmid-mediated resistance or is the mass use of antimicrobials responsible for the selection of resistant strains. These facts clearly highlight the need for new novel therapeutic agents have become necessary (Stephanie et al. 2021).
Malla et al. (2014) have been isolated 98 Vibrio cholerae from 877 stool specimens and recorded the 11.17% of the isolates were isolated. They used the biochemical tests: IMViC, triple sugar iron test, oxidation/fermentation test, Urease test, Nitrate test to identify the isolates and the Vibrio cholerae isolates were identified as belonging to the serogroup 01. E1 Tor biotype and Ogawa serotype (Malla et al. 2014; Stephanie et al. 2021).
But in the present investigation has isolated 50 Vibrio cholerae from 100 stool samples. Similar biochemical tests were used. According to the Guidance of WHO the Vibrio cholerae 01 Serogroup was identified in this study. Adeleye et al. (2010) have been isolated Vibrio spp. from the seafood, 10 antibiotics were used for antimicrobial susceptibility test by disc diffusion method on Mueller Hinton Agar. Plasmid DNA extraction was done using the alkaline lysis method of the Birnboim and Dolly method. Only four of the isolates move plasmid DNA, V. alginolyticus carried plasmids of molecular weight 25.00 kbp and 9.416 kbp each.
Two other isolates of V. mimicus harboured very small plasmids of molecular weight 4.361kbp each. 10 antibiotic discs were used for the antimicrobial susceptibility test; the dissimilar method was used in this test. The extraction of plasmid was done by the method of boiling techniques (Holmes and Quigley 1981). In this study above 50%, resistant strains were selected for plasmid extraction. All the above 50% resistant two isolates were showed plasmids born both are were harbored one fragment of molecular weight like 1500bp respectively (Stephanie et al. 2021).
A total of 250 different samples from poultry and poultry environments were examined and a total of 24 (9.6%) samples were detected as positive for Vibrio cholerae. Out of a total of 50 samples 8, 12, 3 and 1 samples were detected positive for V. cholerae respectively in the cloacal swab, intestinal fluid, egg surface and hand wash of the chicken handlers. The highest positive samples 24% for V. cholerae detection was found in the case of intestinal fluid followed by the samples of the cloacal swab (16%) and egg surface (6%) and the hand wash of chicken handlers was the lowest.
Antibiotic susceptibility pattern of V. cholerae has been conducted against selected 11 antibiotics and agar plates, All the 15 isolates of V. cholerae (designated as Vc1, Vc2, Vc3 and so on up to Vc15) were found to the 100% resistant to Erythromycin, Ampicillin, Kanamycin, Penicillin, Tetracycline, and Rifampicin and 60% of the strain resistant to the Cephalexin, Streptomycin (Hamasalih et al. 2020).
100% cases of tested strains were found sensitive to Norfloxacin, Gentamycin, and Neomycin (Sule et al. 2019). Similarly in the present investigation, 10 antibiotics were used for antibiotic susceptibility patterns. The highest percentage of resistance was shown in Erythromycin (97%) and the highest susceptibility was shown in Erythromycin (96%) followed by ampicillin and ticarcillin (90%) (Sule et al. 2019).
CONCLUSION
The findings of the present study has indicated that Vibrio cholerae strains were found to develop resistance day by day to the currently used antibiotics. Presence of a large number of Multi-Drug Resistant isolates it makes necessitates curbing irrational use of antibiotics to prevent further spread of drug resistance. This example needs some novel therapeutic drugs in future to beat this sort of dreadful disease.
ACKNOWLEDGEMENTS
This study was financially supported by the Chikkanna Government Arts College.
Conflict of interests: Authors declare that have no conflicts of interests to disclose.
REFERENCES
Adeleye A., Vivian E., Rita N. et al. (2010). Antimicrobial susceptibility of potentially pathogenic halophilic Vibrio species isolated from seafoods in Lagos, Nigeria. African Journal of Biotechnology 7 (20): 3791-3794.
Canellas A.L.B., Lopes I.R., Mello M.P. et al. (2021). Vibrio species in an Urban Tropical Estuary: Antimicrobial Susceptibility, Interaction with Environmental Parameters, and Possible Public Health Outcomes. Journals of Microorganisms .9 (5) 1-18 https://doi.org/ 10.3390/microorganisms9051007.
Das B., Verma J., Kumar P. et al. (2020). Antibiotic resistance in Vibrio cholera understanding the ecology of resistance genes and mechanisms. Science direct 38 A83–A92.
Garberna S.C., Chub T.C., Yangc P. et al. (2021). Clinical and socio-environmental determinants of multidrug-resistant Vibrio cholerae 01 in older children and adults in Bangladesh. International Journal of Infectious Diseases. 105 436–441.
Hamasalih R.M., Hassan A.N. and Saeed R.H. (2020). Plasmid profile and transformation efficiency in Staphylococcus aureus isolated from different clinical specimens. Pakistan Journal of Medical and Health Sciences. 14 (3): 1584-1593.
Holmes D and Quigley M (1981). A rapid boiling method for the preparation of bacterial plasmids. Anal. Biochem. 114: 193-197.
Lonappan S., Golecha R. and Nair G.B. (2019). Contrasts, contradictions and control of cholera. Science Direct. 38: A4–A6.
Malla S., Dumre S.P., Shakya G. et al. (2014). The challenges and successes of implementing a sustainable antimicrobial resistance surveillance programme in Nepal. BMC Public Health. 14 (269): 1-7.
Murphy S.G., Johnson B.A., Ledoux C.M. et al. (2021). Vibrio cholerae’s mysterious Seventh Pandemic Island (VSP-II) encodes novel Zur-regulated zinc starvation genes involved in chemotaxis and cell congregation. PLoS Genet 17 (6): 1-31. e1009624 https://doi.org/10.1371/journal.pgen.1009624.
Sule A., Nkene I. and Ekeme K.I. (2019). Antibiotic Resistance and Plasmid Profile of Escherichia Coli from Feacal Samples of Cattle From Abattoir In Keffi Metropolis, Nasarawa State Nigeria. International Journal of Scientific and Research Publications. 9 (9): 343-349.
Talukder B., Chakma F., Sabuj A.A.M. et al. (2019). Identification, prevalence and antimicrobial susceptibility of major pathogenic bacteria in exportable shrimp (Penaeus monodon) of Southern Bangladesh. Int J Fish Aquat Stud. 7 (1): 96-9.
Uddin M.E., Akter T., Sultana P. et al. (2018). Isolation, identification and antimicrobial susceptibility profile analysis of Vibrio cholerae O1 from stool samples of Bangladesh. Advances in Microbiology. 8:188-196.


