1Research Scholar, School of Life Sciences, Jaipur National University, Jaipur, 302017, India.
2Associate Professor, School of Life Sciences, Jaipur National University, Jaipur, 302017, India.
3Vice Chancellor, Shobhit Institute of Engineering & Technology (Deemed to be University), Meerut, 250110, India.
Corresponding author email: amarprakashgarg@yahoo.com
Article Publishing History
Received: 10/02/2020
Accepted After Revision: 20/03/2020
The first thick milk produced immediately after the delivery is called human colostrum (HC). Its composition and functions are quite different than mature milk. It contains high levels of proteins, vitamins, immunoglobulins, carbohydrates, amino acids and many other nutrients. Apart from its nutritional aspects, HC also contains large number of Lactic Acid Bacteria (LAB) with huge probiotic potential. These LAB helps in nourishment, proper growth and development of infants in the early stages of life. The main objective of the study was to characterize and evaluate the probiotic potential of LAB from HC. The study showed several LAB with probiotic potential. The isolated LAB fulfilled all the necessary criteria of a standard probiotics such as growth at low pH, different temperatures, tolerance against bile salts, resistance against antibiotics and antimicrobial activities against common human pathogens. Four isolates of the study were found to be very promising in showing resistance against antibiotics and antimicrobial response against common pathogens such as Escheria coli ATCC 25922, Proteus vulgaris ATCC 33420, Staphylococcus aureus ATCC 25922, Salmonella typhi ATCC 733 and Pseudomonas aeruginosa ATCC 27853. On the basis of biochemical characterization, the isolates were identified as Lactobacillus brevis, L. acetotolerans, L. casei and Pediococcus acidilactici. The present paper deals with the isolation, characterization and evaluation of probiotic potentials of LAB isolated from HC.
Antagonistic activity, Human Colostrum, Infant Gut, Lactic Acid Bacteria, Probiotics.
Arya R, Singh J, Garg A. P. Isolation, Characterization and Evaluation of Probiotic Potential of Lactic Acid Bacteria Isolated from Human Colostrum. Biosc.Biotech.Res.Comm. 2020;13(1).
Arya R, Singh J, Garg A. P. Isolation, Characterization and Evaluation of Probiotic Potential of Lactic Acid Bacteria Isolated from Human Colostrum. Biosc.Biotech.Res.Comm. 2020;13(1). Available from: https://bit.ly/36qMxrd
Copyright © Arya et al., This is an open access article distributed under the terms of the Creative Commons Attribution License (CC-BY) https://creativecommons.org/licenses/by/4.0/, which permits unrestricted use distribution and reproduction in any medium, provide the original author and source are credited.
INTRODUCTION
For many years, HC was considered to be a sterile fluid, but recent studies have revised this dogma (Fernandez et al., 2013). The period of flow of HC is from 1st to 6th day of lactation. The milk produced after the 6th day is mature milk (Castellote et al., 2011). HC is a thick fluid rich in nutrients and contains vitamins, proteins, amino acids, carbohydrates and lipids along with several immune cells which provide immunity to infants in early stages of growth and development (Ballard et al., 2013). Recent studies reveals that apart from all the nutritional aspects of HC, it also contains large number of probiotic bacteria which helps in digestion and protection against infections (Marchesi et al., 2016). The study on milk of Rheses monkey (Mucaca mulatta) first showed that milk contains 19 different species of bacteria belonging to 8 genera in its constituents (Jin et al., 2011). HC also contains large number of other bacteria (Hunt et al., 2011). These bacteria also play a very important role in the development of immune system of infant (Wang et al., 2018).
The number of bacteria in HC are about thrice more than mature milk. The number of bacteria in mature milk lower downs with the continuous regular flow of milk (McGuire et al., 2015). From the studies carried out in the past, majority of the bacteria isolated from human milk were generally Lactic Acid Bacteria (LAB) (Jost et al., 2015). LAB is a large group of bacteria used worldwide as a probiotic. This group of bacteria involves the microorganisms of genera Lactobacillus, Lactococcus, Aerococcus, Enterococcus, Pediococcus, Leuconostoc, Streptococcus, Sporolactobacillus, Vagococcus and Carnobacterium (Pavli et al., 2018 & Neha 2019).
The bacteria of these genera have high probiotic potential and have been proven safe for human consumption (Guesh et al., 2019). The first bacteria that enters the infant gut is from HC. These bacteria enters infant gut through HC and remains in the gut for entire life (Pang et al., 2007). The gut microflora get involves in various biochemical processes and serves several functions in the welfare of human gut (Dunlop et al., 2015). LAB have innumerable health benefits such as blood pressure lowering (Robles-Vera et al., 2017), prevention of colon cancer (Rafter 2003), reduction of allergic symptoms (Cuello-Garcia et al., 2017), reduction of cholesterol (Agerholm-Larsen et al., 2002), boosting of immune system (King et al., 2014), prevention of urinogenital infections (Shortliffe et al., 2013), reduction of Helicobacter pylori infections (Hamilton 2003), Intestinal Inflammation (Jin-Sil et al., 2018), antimicrobial effects on pathogens (Tankoano et al., 2019) and many more. Therefore, it can also be said that LAB are boon to infant gut. The present study deals with isolation, characterization and evaluation of probiotic potential of HC.
MATERIAL AND METHODS
Sample Collection: Total 60 different HC samples were collected from lactating mothers who voluntarily gave their consent for our study. All the samples were collected immediately after the delivery from the maternity ward of Jaipur National University Hospital, Jaipur (India). The tubes used for sample collection were autoclaved using standard protocols. The nipples of lactating mothers were cleaned properly with cotton dipped in alcohol to avoid any contaminations of breast skin microflora. The mid flow of HC was carefully aseptically collected in the tubes with the help of experts.
Isolation of LAB:The isolation of LAB from HC was quickly processed after completion of sample collection. The HC samples were serially diluted upto 10-6 using sterile peptone water. The last three dilutions were inoculated on MRS agar plates using Spread Plate Technique. The inoculated plates were incubated at 37℃ for 48 h under anaerobic conditions using anaerobic gas jar.
Enumeration of LAB:After incubation, the bacterial colonies were counted using digital colony counter and Colony Forming Unit (CFU) per mL of HC were calculated using standard method.
Biochemical Characterization of Isolates:Isolated bacteria were sub-cultured to get pure form of colony. The colony characteristics of each isolate were recorded carefully. The pure colonies were further chosen for biochemical characterization. Gram’s staining, Catalase test, Oxidase Test, Arginine Hydrolysis Test and Sugar Fermentation tests were performed to characterize the isolates to be LAB as per recommendation of Bergey’s Manual of Determination Bacteriology.
Gram’s Staining:A single drop of sterile water was dropped on a clean glass slide and a pure colony of isolate was picked from the plate and was mixed gently to prepare a smear. The smear was heat fixed carefully. The standard procedure of Gram’s staining was followed and the slide was observed under oil emulsion lens (10x X 100x) of compound microscope. As LAB are Gram positive in nature, all the isolates which showed Gram positive nature were further processed for other biochemical tests.
Catalase Test:Catalase test was performed for all the isolates which were Gram’s positive. Catalase is a type of enzyme which is produced by several microorganisms that breaks down hydrogen peroxide into water and oxygen and forms bubbles of gas. The 3% hydrogen peroxide solution was mixed gently on the surface of clean glass slide and was observed for bubble formation. As LAB are catalase negative, all the isolates that showed negative results of catalase test were further tested for oxidase test.
Oxidase Test:All the isolates which showed catalase activity negative were further tested for oxidase test. Cytochrome c oxidase is an enzyme found in several bacterial electron transport chain. Presence of cytochrome c oxidase oxidizes the reagent called tetramethyl-phenylenediamine into indophenols (purple color) end product. As LAB are oxidase negative, all the isolates which showed negative results were further tested for its arginine hydrolysis.
Arginine Hydrolysis Test:Nessler’s reagent and arginine MRS medium were used to check the production of ammonia from arginine. 5 mL of MRS broth was transferred to empty test tube and 100 μl of test culture (O. D 1.0 at 600 nm) was inoculated and the tubes were incubated at 37±1°C for 24h. After incubation, an equal volume of Nessler’s reagent was added to each tube. The immediate appearance of dark orange color was interpreted as positive (presence of ammonia) while indication of yellowish color was interpreted as negative reaction (absence of ammonia) (Kavitha and Devasena 2013).
Sugar Fermentation Test:Carbohydrate when fermented by microorganisms form an acid or acid with gas at the end. Depending on the microorganisms involved, the end products may vary. All the isolates which were Gram positive and catalase and oxidase negative were tested for their sugar fermentation activity. Sugars were prepared using standard protocol (HiMedia) and each tube of sugar contained Durham’s tube in inverted position. Each isolate was inoculated in all different sugars (Glucose, Lactose, Maltose, Fructose, Mannitol, Galactose and Sucrose) to note down the breakdown of sugars into acid and/or acid + gas. Incubation for 48 h at 37 ℃ were given to all the sugars. Results were recorded after completion of incubation period. On the basis of Sugar fermentation activity, the isolates were identified using Bergey’s Manual of Systematic Bacteriology (Hammes P et al., 2009).
Determination of Probiotic Potential.After biochemical characterization, all the isolates were tested for their probiotic potential by testing their growth at low pH, different temperatures, tolerance against bile salts, resistance against common human pathogens and resistance to antibiotics.
Growth at low pH:The pH of human stomach ranges between 2 to 3. It is also believed that food eaten by us stays in stomach for at least 4 h (Bistha N et al., 2019). Therefore, it is the necessary for the isolate to survive at low pH for more than 4 h. To check the growth of isolates at low pH, all the isolates were inoculated in peptone water prepared with different pH (6, 5, 4, 3, 2) for a period of 6 h. After incubation period, the isolates were inoculated on MRS agar plates and were incubated under anaerobic conditions to check their survival at different pH. All the isolates were further checked for their tolerance against bile salts.
Tolerance against Bile Salts:The concentration of bile salts in the intestine is believed to be 0.3% (w/v) and the food eaten stays in small intestine is suggested to be 4 h (Kumari A et al., 2019). Therefore, all the isolates were examined for their growth at different bile salts concentrations. Peptone water with different bile salts concentration was prepared using Oxoid and active cultures of isolates were inoculated in the medium for 6 h. After incubation, the isolates were inoculated on MRS agar plates for its viable count. The isolates which showed good growth on plates were further tested for their growth at different temperatures.
Growth at Different Temperatures: To examine the growth of isolates at different temperature, the active cultures of isolates were inoculated on MRS agar plates and were incubated at different temperatures (25, 30, 35, 40 ℃) in anaerobic conditions. The isolates which showed best growth at both high as well as low temperatures were further screened for their resistance against antibiotics.
Resistance to Antibiotics:The isolates which gave best growth at high as well as low temperature were tested for their resistance against common antibiotics using Kirby Bauer method. The isolates were spreaded on the entire surface of MH agar plates and the discs of antibiotics with different concentrations were placed on the surface of agar and gently pressed. The plates were allowed to incubate at room temperature for 24-48 h. The isolates which did not give appropriate zone of inhibition around the discs of antibiotics according to standard chart were further examined for their antimicrobial activities against common human pathogens.
Antimicrobial activity:All the isolates which fulfilled the above mentioned criteria were further tested for their antimicrobial activities against common human pathogens using agar well diffusion method. The indicator pathogenic microorganisms were spreaded on the entire surface of Muller Hilton (MH) agar plates and using a sterile core borer of 7 mm diameter. 5 different wells of same size were made by puncturing the MH agar plates. Using micropipettes, 80 μL of overnight grown culture of isolate were inoculated carefully in the wells. The plates were incubated for 24 h in upright position. Thereafter, the zone of inhibition were measured. The isolates which showed greater zones of inhibition were considered having good probiotic potential.
RESULTS AND DISCUSSION
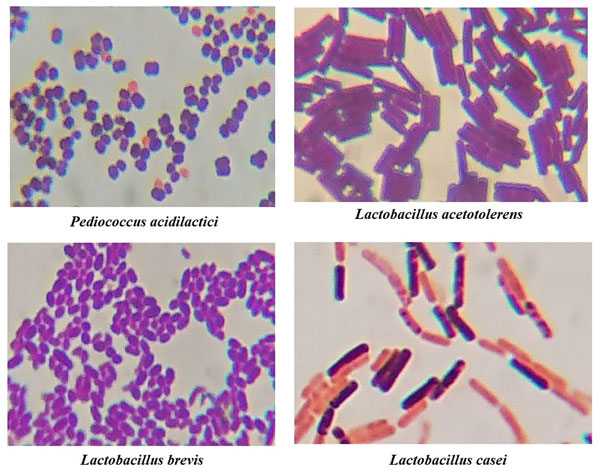
A total of 130 LAB were isolated from the HC of 60 different lactating mothers. The isolates were identified on the basis of physiological and biochemical characteristics. On the basis of Bergey’s Manual of Systematic Bacteriology, 72 different species of LAB were identified. Of these, 4 isolates of LAB were found to be very promising with the potential of probiotics. These isolates were selected on the basis of their antimicrobial activities and their resistance against antibiotics. The average number of LAB count per ml of HC of a health lactating mothers were found to be 108 to 109. The LAB count was measured on the basis Standard Plate Count (Total Viable Count). The isolates were initially confirmed by using biochemical test such as Catalase, Oxidase, Grams Staining, Arginine Hydrolysis test and Sugar Fermentation test. All the isolates in the present study were found Gram’s positive, Catalase and Oxidase negative and also had the capacity to breakdown sugars into acids and gas [Table 1]. On the basis of their Sugar Fermentation activity and Gram’s morphology [Figure 1], the isolates were identified using Bergey’s Manual of Systematic Bacteriology (Hammes et al., 2009).
Figure 1: Microscopic observation of Gram’s Staining
Determination of LAB to be potentially probiotic: All the isolates identified as LAB through biochemical tests were further screened for determining their probiotic potential. Firstly, the growth of isolates were checked at low pH. Out of 130 isolates, 79 showed its positive growth at pH 2 which were further screened for their tolerance against different bile salts concentrations. Out of 130 total isolates, 96 were found to be prominent against tolerating the 0.3% (w/v) bile salts concentrations. These isolates were further examined for their growth at different temperatures. Out of 130 isolates, 77 showed a good growth at 40 ℃ and even at 25 ℃. On basis of these three criteria’s, 34 best isolates were selected for checking their resistance against antibiotics from which 16 best isolates were screened for testing their antimicrobial activity against common human pathogens. Out of 16 isolates, only 4 showed very high degree of zone of inhibition against pathogenic bacteria. The details of these for isolates are mentioned below in Table I, II, III and IV.
Table 1 :Fermentation of different sugars for identification of isolates as per the recommendations of Bergey’s Manual
| Sample No. | Isolate no. | Glucose | Maltose | Lactose | Mannitol | Galactose | Fructose | Sucrose | Identification Based on Bergey’s Manual | |||||||
| A | G | A | G | A | G | A | G | A | G | A | G | A | G | |||
| HC1 | I1001 | + | + | + | + | + | + | – | – | + | + | + | – | + | – | Lactobacillus acidophilus |
| I1002 | + | + | + | + | + | – | – | – | + | – | + | – | + | – | Lactobacillus sakei | |
| HC2 | I2001 | + | – | + | + | + | – | + | – | + | – | + | – | +` | – | Lactobacillus paracasei |
| I2002 | + | + | + | – | + | – | – | – | + | – | + | + | + | + | Lactobacillus fermentum | |
| I2003 | + | + | + | + | + | – | + | – | + | – | + | – | + | – | Streptococcus oralis | |
| HC3 | I3001 | + | + | + | + | + | + | – | – | + | + | + | – | + | – | Lactobacillus gasseri |
| I3002 | + | + | + | – | + | – | + | – | + | – | + | – | – | – | Lactobacillus agilis | |
| HC4 | I4001 | + | + | + | – | + | – | + | – | + | – | + | – | + | – | Bifidobacterium longum |
| I4002 | + | – | + | + | + | – | – | – | + | – | + | – | +` | – | Lactobacillus rhamnosus | |
| HC5 | I5001 | + | + | + | + | + | + | + | – | + | + | + | – | + | – | Bifidobacterium breve |
| I5002 | + | + | + | + | + | – | + | – | + | + | + | + | – | – | Pediococcus demnosus | |
| HC6 | I6001 | + | + | + | + | – | – | + | – | + | – | + | + | + | – | Lactobacillus oris |
| I6002 | + | + | + | – | + | – | + | – | – | – | – | – | + | – | Lactobacillus curtus | |
| HC7 | I7001 | + | + | + | – | + | + | + | – | + | + | + | + | + | – | Bifidobacterium magnum |
| I7002 | + | + | + | + | + | – | – | – | + | + | + | + | + | – | Lactococcus garvieae | |
| HC8 | I8001 | + | + | + | + | – | – | + | + | + | + | + | – | – | – | Bifidobacterium dentium |
| I8002 | + | + | + | + | + | + | – | – | + | + | – | – | + | – | Lactobacillus johnsonii | |
| HC9 | I9001 | + | + | + | + | + | + | + | – | – | – | + | – | + | + | Lactobacillus gasseri |
| I9002 | – | – | + | + | + | – | + | – | + | – | – | – | – | – | Lactobacillus helviticus | |
| I9003 | + | – | + | + | + | – | + | – | + | – | + | – | +` | – | Lactobacillus backii | |
| HC10 | I0101 | + | + | + | + | + | – | + | – | + | – | + | – | + | – | Lactobacillus parakefiri |
| I0102 | + | – | + | – | + | + | – | – | + | – | + | – | + | – | Stretococcus bovis | |
| HC11 | I1101 | + | + | + | + | – | – | + | – | + | – | + | – | – | – | Lactobacillus silage |
| I1102 | + | + | + | + | + | – | + | – | + | – | – | – | + | – | Lactobacillus rennini | |
| HC12 | I1201 | + | + | + | + | + | – | + | – | + | – | + | – | + | – | Bifidobacterium bifidum |
| I1202 | + | – | + | – | + | – | + | + | + | + | + | – | + | – | Lactobacillus rapi | |
| HC13 | I1301 | + | + | + | + | + | + | + | + | + | + | + | + | + | – | Pediococcus cellicola |
| I1302 | + | + | + | + | + | – | – | – | – | – | + | – | – | – | Lactobacillus ozensis | |
| HC14 | I1401 | + | + | – | – | – | – | + | – | + | – | + | – | + | + | Lactobacillus helviticus |
| I1402 | + | – | + | + | + | + | – | – | + | – | + | – | – | – | Bifidobacterium hapal | |
| HC15 | I1501 | – | – | + | + | + | – | + | – | – | – | + | – | + | – | Bifidobacterium merycicum |
| I1502 | + | + | + | + | + | + | – | – | + | + | + | – | + | – | Lactobacillus acidipiscis | |
| I1503 | + | + | + | + | + | – | + | – | + | – | + | – | + | – | Lactococcus piscium | |
| HC16 | I1601 | + | + | + | + | + | + | + | + | + | – | + | + | + | – | Lactobacillus acetotolerens |
| I1602 | + | + | + | + | + | + | + | – | + | – | – | – | + | Lactobacillus florum | ||
| HC17 | I1701 | + | + | + | + | + | + | + | – | + | + | + | – | + | – | Bifidobacterium breve |
| I1702 | + | + | + | + | + | – | + | – | + | – | + | + | – | – | Lactococcus plantarum | |
| HC18 | I1801 | + | + | + | + | + | – | + | – | + | – | + | – | + | – | Bifidobacterium reuteri |
| I1802 | + | + | + | – | + | + | + | – | + | – | + | + | – | – | Bifidobacterium bifidum | |
| HC19 | I1901 | + | + | + | – | + | – | – | – | + | – | + | + | + | + | Lactobacillus plantarum |
| I1902 | + | + | + | + | + | + | + | – | + | + | + | – | + | – | Lactobacillus acidophilus | |
| HC20 | I0201 | + | + | + | + | + | – | + | – | + | – | + | – | – | – | Lactobacillus agalis |
| I0202 | + | + | + | + | – | – | + | + | + | + | + | – | – | – | Bifidobacterium dentium | |
| HC21 | I2101 | – | – | + | – | + | + | + | – | + | + | + | – | + | – | Pediococcus inopinatus |
| I2102 | + | + | + | – | + | + | + | + | – | – | + | – | + | – | Lactobacillus casei | |
| I2103 | + | + | + | + | + | + | – | – | + | – | + | + | + | – | Lactobacillus larvae | |
| HC22 | I2201 | + | – | + | + | – | – | + | – | + | – | – | – | + | – | Lactobacillus nagelii |
| I2202 | + | + | + | + | + | + | + | + | – | – | + | – | + | – | Lactococcus formosensis | |
| HC23 | I2301 | + | + | + | – | + | – | + | – | – | – | – | – | + | – | Pediococcus stilesii |
| I2302 | – | – | + | + | + | – | + | – | + | – | – | – | – | – | Lactobacillus helviticus | |
| HC24 | I2401 | + | + | + | + | + | – | – | – | + | + | + | + | + | – | Lactobacillus brevis |
| I2402 | + | – | + | + | + | – | + | – | + | – | + | – | +` | – | Lactobacillus paracasei | |
| HC25 | I2501 | + | + | + | + | + | + | – | – | + | + | – | – | + | – | Pediococcus ethanoliduran |
| I2502 | + | + | + | – | + | – | + | – | + | – | + | – | + | – | Lactobacillus pontis | |
| I2503 | – | – | + | + | + | – | + | – | + | – | – | – | – | – | Lactococcus raffinolactis | |
| HC26 | I2601 | + | + | – | – | + | + | + | – | + | + | + | + | + | – | Lactobacillus nuruki |
| I2602 | + | + | + | + | + | – | + | – | + | – | + | – | + | – | Bifidobacterium reuteri | |
| HC27 | I2701 | + | – | + | – | + | + | – | – | + | – | + | – | + | – | Bifidobacterium bombi |
| I2702 | + | + | + | + | – | – | + | – | + | – | + | – | – | – | Lactobacillus perolens | |
| HC28 | I2801 | + | + | + | + | + | – | + | – | + | – | – | – | + | – | Pediococcus pentosaceus |
| I2802 | + | + | + | + | + | – | + | – | + | – | + | – | + | – | Lactobacillus raoutii | |
| HC29 | I2901 | – | – | + | + | + | – | + | – | + | – | – | – | – | – | Lactobacillus helviticus |
| I2902 | + | + | + | + | + | + | + | + | + | + | + | + | + | – | Lactobacillus mobilis | |
| HC30 | I0301 | + | + | + | + | + | – | – | – | – | – | + | – | – | – | Streptococcus ferus |
| I0302 | + | – | + | + | + | – | + | – | + | – | + | – | +` | – | Lactobacillus buchnerii | |
| HC31 | I3101 | + | + | + | + | + | – | + | – | + | – | + | – | + | – | Lactobacillus parakefiri |
| I3102 | – | – | + | + | + | – | + | – | – | – | + | – | + | – | Lactococcus hircilactis | |
| I3103 | + | + | + | + | + | – | + | – | + | – | + | – | – | – | Lactobacillus agalis | |
| HC32 | I3201 | + | + | + | + | + | – | + | – | + | – | + | – | + | – | Lactobacillus casei |
| I3202 | + | + | + | + | + | – | + | – | + | – | + | – | + | – | Bifidobacterium reuteri | |
| HC33 | I3301 | + | + | + | – | + | + | + | – | + | – | + | + | – | – | Bifidobacterium bifidum |
| I3302 | + | + | – | – | + | – | + | – | + | – | + | – | + | – | Pediococcus claussenii | |
| HC34 | I3401 | + | + | + | + | + | – | + | – | + | – | + | + | – | – | Aerococcus suis |
| I3402 | + | + | + | – | – | – | + | – | + | – | + | + | + | – | Bifidobacterium boum | |
| HC35 | I3501 | + | + | + | + | + | – | + | – | + | – | + | – | + | – | Lactobacillus pasteurii |
| I3502 | + | + | + | + | + | – | + | – | + | – | + | – | + | – | Lactobacillus raoutii | |
| HC36 | I3601 | + | + | + | – | + | – | – | – | + | – | + | + | + | + | Lactococcus laundensis |
| I3602 | + | + | + | + | + | – | + | – | + | – | + | – | + | – | Lactobacillus saniviri | |
| HC37 | I3701 | + | + | + | + | + | – | + | – | + | – | + | – | + | – | Bifidobacterium reuteri |
| I3702 | + | + | + | – | + | – | + | – | + | – | + | – | + | – | Lactobacillus rogosae | |
| HC38 | I3801 | + | + | + | + | – | – | + | + | + | + | + | – | – | – | Bifidobacterium dentium |
| I3802 | + | – | + | + | + | – | – | – | + | – | + | – | +` | – | Lactobacillus sunkii | |
| I3803 | + | + | + | + | + | – | + | – | + | – | + | + | – | – | Streptococcus downei | |
| HC39 | I3901 | + | + | + | + | + | + | + | – | + | – | – | – | + | Lactobacillus florum | |
| I3902 | + | – | + | + | + | – | + | – | + | – | + | – | +` | – | Bifidobacterium myosotis | |
| HC40 | I0401 | + | + | + | + | + | – | + | – | + | – | + | – | + | – | Lactobacillus sakei |
| I0402 | + | + | + | + | + | + | – | – | + | + | + | – | + | – | Pediococcus acidilactici | |
| HC41 | I4101 | + | + | + | – | + | + | + | + | – | – | + | – | + | – | Lactobacillus casei |
| I4102 | + | + | + | + | + | + | + | – | + | + | + | – | + | – | Lactobacillus gasseri | |
| HC42 | I4201 | + | + | + | – | + | + | + | – | + | – | + | + | – | – | Bifidobacterium bifidum |
| I4202 | + | – | + | + | – | – | + | – | + | – | – | – | + | – | Lactobacillus acetotoleren | |
| HC43 | I4301 | + | + | + | + | + | – | + | – | + | – | – | – | + | – | Lactobacillus rennini |
| I4302 | + | + | + | + | + | – | + | – | + | – | + | – | + | – | Lactococcus piscium | |
| HC44 | I4401 | + | + | + | – | + | – | – | – | + | – | + | + | + | + | Lactobacillus plantarum |
| I4402 | + | + | + | – | + | + | + | + | – | – | + | – | + | – | Lactobacillus ozensis | |
| HC45 | I4501 | + | + | + | + | + | + | + | + | + | + | + | + | + | – | Pediococcus cellicola |
| I4502 | + | + | + | + | + | + | + | – | + | + | + | – | + | – | Lactobacillus acidophilus | |
| I4503 | + | + | + | – | + | – | – | – | + | – | + | + | + | + | Lactobacillus fructivorans | |
| HC46 | I4601 | + | + | + | + | + | – | + | – | + | – | + | + | – | – | Pediococcus parvulus |
| I4602 | + | + | + | + | + | – | + | – | + | – | + | – | + | – | Lactobacillus pasteurii | |
| HC47 | I4701 | – | – | + | + | + | – | + | – | – | – | + | – | + | – | Lactococcus hircilactis |
| I4702 | + | + | + | + | + | – | + | – | + | + | + | + | – | – | Pediococcus demnosus | |
| HC48 | I4801 | + | + | + | + | – | – | + | – | + | – | + | + | + | – | Lactobacillus oris |
| I4802 | + | – | + | + | – | – | + | – | + | – | – | – | + | – | Lactobacillus acetotoleren | |
| HC49 | I4901 | + | + | + | + | + | + | – | – | + | + | + | – | + | – | Pediococcus acidilactici |
| I4902 | + | + | + | + | + | – | + | – | + | – | + | – | + | – | Bifidobacterium reuteri | |
| HC50 | I0501 | + | + | + | + | + | – | – | – | + | + | + | + | + | – | Lactococcus garvieae |
| I0502 | + | + | + | + | – | – | + | – | + | – | + | – | – | – | Lactobacillus perolens | |
| HC51 | I5101 | + | + | + | + | + | – | + | – | + | – | + | – | + | – | Lactobacillus pasteurii |
| I5102 | + | + | + | + | + | – | + | – | + | – | + | – | + | – | Bifidobacterium bifidum | |
| HC52 | I5201 | + | + | + | + | – | – | + | – | + | – | + | – | – | – | Lactobacillus perolens |
| I5202 | + | + | + | – | + | + | + | – | + | – | + | + | – | – | Aerococcus sanguinicola | |
| I5203 | + | + | + | – | + | – | – | – | + | – | + | + | + | + | Lactobacillus plantarum | |
| HC53 | I5301 | + | + | + | + | + | + | + | – | + | + | + | – | + | – | Lactobacillus gasseri |
| I5302 | + | + | + | + | + | – | + | – | + | – | + | – | – | – | Lactobacillus vini | |
| HC54 | I5401 | + | + | + | + | + | + | – | – | + | – | + | + | + | – | Lactobacillus larvae |
| I5402 | + | – | + | + | + | – | + | – | + | – | + | – | +` | – | Lactobacillus paracasei | |
| HC55 | I5501 | – | – | + | + | + | – | + | – | – | – | + | – | + | – | Lactococcus hircilactis |
| I5502 | + | + | + | + | + | + | + | – | – | – | + | – | + | + | Lactobacillus gasseri | |
| HC56 | I5601 | + | – | + | + | – | – | + | – | + | – | – | – | + | – | Lactobacillus acetotoleren |
| I5602 | + | + | + | + | + | + | + | + | + | + | + | + | + | – | Lactobacillus mobilis | |
| HC57 | I5701 | + | + | + | + | + | – | + | – | + | – | + | – | – | – | Lactobacillus agalis |
| I5702 | + | + | + | + | + | – | + | – | + | – | + | – | + | – | Lactococcus lactis | |
| I5703 | + | + | + | – | + | + | + | – | + | – | + | + | – | – | Bifidobacterium minimum | |
| HC58 | I5801 | + | + | + | + | – | – | + | – | + | – | + | + | + | – | Lactobacillus oris |
| I5802 | + | + | + | + | + | – | + | – | + | – | + | – | + | – | Lactobacillus casei | |
| HC59 | I5901 | + | + | + | – | + | – | + | – | – | – | – | – | + | – | Pediococcus stilesii |
| I5902 | + | + | + | – | + | – | + | – | + | – | + | – | + | – | Bifidobacterium longum | |
| HC60 | I0601 | + | + | + | + | + | + | + | + | + | + | + | + | + | – | Lactobacillus mobilis |
| I0602 | + | + | + | + | + | – | – | – | + | + | + | + | + | – | Lactococcus garvieae | |
Table 2: Determination of probiotic potential based on growth at low pH, bile salt tolerance and growth at variable temperatures
| Sample No. | Isolate no. | Growth at different pH | Bile Salt Tolerance (%) | Growth at different Temperatures (℃) | ||||||||||
| pH6 | pH5 | pH4 | pH3 | pH2 | 0.2 | 0.3 | 0.4 | 0.5 | 25 | 30 | 35 | 40 | ||
| HC1 | I1001 | + | + | + | – | – | + | + | + | – | + | + | + | + |
| I1002 | + | + | + | + | + | + | + | + | + | – | + | + | + | |
| HC2 | I2001 | + | + | + | + | + | + | – | – | – | + | + | + | + |
| I2002 | + | + | + | + | + | + | + | + | – | – | – | + | + | |
| I2003 | + | + | + | – | – | + | + | + | + | + | + | + | – | |
| HC3 | I3001 | + | + | + | + | + | – | – | – | – | + | + | + | + |
| I3002 | + | + | – | – | – | + | + | + | + | – | + | + | + | |
| HC4 | I4001 | + | + | + | + | + | + | – | – | – | + | + | + | + |
| I4002 | + | + | + | + | – | + | + | + | + | + | + | + | – | |
| HC5 | I5001 | + | + | + | + | – | + | + | + | + | + | + | + | + |
| I5002 | + | + | + | + | + | – | – | – | – | – | – | + | + | |
| HC6 | I6001 | + | + | – | – | – | + | + | + | + | + | + | + | – |
| I6002 | + | + | + | + | + | + | – | – | – | + | + | + | + | |
| HC7 | I7001 | + | + | + | – | – | + | + | + | + | + | + | + | + |
| I7002 | + | + | + | + | + | + | + | + | + | + | + | + | + | |
| HC8 | I8001 | + | + | + | + | + | + | – | – | – | + | + | + | + |
| I8002 | + | + | + | + | + | + | + | + | + | – | + | + | + | |
| HC9 | I9001 | + | + | + | + | + | + | + | + | + | + | + | + | + |
| I9002 | + | + | + | – | – | + | + | + | + | + | + | + | – | |
| I9003 | + | + | + | + | – | – | – | – | – | + | + | + | + | |
| HC10 | I0101 | + | + | + | + | + | + | + | + | + | – | – | + | + |
| I0102 | + | + | + | + | + | + | + | + | + | + | + | + | + | |
| HC11 | I1101 | + | + | + | + | – | + | + | + | + | + | + | + | + |
| I1102 | + | + | + | + | + | + | + | – | – | – | + | + | + | |
| HC12 | I1201 | + | + | + | – | – | + | + | + | + | + | + | + | + |
| I1202 | + | + | + | + | + | – | – | – | – | + | + | + | + | |
| HC13 | I1301 | + | + | + | + | + | + | + | + | + | + | + | + | + |
| I1302 | + | + | + | + | + | + | + | + | + | – | + | + | + | |
| HC14 | I1401 | + | + | – | – | – | + | + | + | + | + | + | + | – |
| I1402 | + | + | + | + | – | – | – | – | – | + | + | + | + | |
| HC15 | I1501 | + | + | + | + | + | + | + | + | + | + | + | + | + |
| I1502 | + | + | + | + | – | + | + | + | + | + | + | + | – | |
| I1503 | + | + | + | + | + | + | + | + | + | – | – | + | + | |
| HC16 | I1601 | + | + | + | + | + | + | + | + | + | + | + | + | + |
| I1602 | + | + | + | – | – | + | – | – | – | + | + | + | + | |
| HC17 | I1701 | + | + | + | + | + | + | + | + | + | + | + | + | + |
| I1702 | + | + | + | + | + | + | + | + | + | – | + | + | + | |
| HC18 | I1801 | + | + | – | – | – | + | + | + | + | + | + | + | + |
| I1802 | + | + | + | + | + | – | – | – | – | + | + | + | + | |
| HC19 | I1901 | + | + | + | + | – | + | + | + | + | + | + | + | + |
| I1902 | + | + | + | + | + | + | + | + | + | – | + | + | + | |
| HC20 | I0201 | + | + | + | + | + | + | – | – | – | + | + | + | + |
| I0202 | + | + | + | + | – | + | + | + | + | + | + | + | – | |
| HC21 | I2101 | + | + | + | + | + | – | – | – | – | + | + | + | + |
| I2102 | + | + | + | – | – | + | + | + | + | + | + | + | + | |
| I2103 | + | + | + | + | + | + | + | + | + | – | – | + | + | |
| HC22 | I2201 | + | + | + | + | + | + | + | + | + | + | + | + | + |
| I2202 | + | + | + | + | + | + | + | – | – | + | + | + | + | |
| HC23 | I2301 | + | + | + | – | – | + | + | + | + | + | + | + | + |
| I2302 | + | + | + | + | + | + | + | + | + | + | + | + | + | |
| HC24 | I2401 | + | + | + | + | + | + | + | + | + | – | – | + | + |
| I2402 | + | + | + | + | + | – | – | – | – | + | + | + | + | |
| HC25 | I2501 | + | + | + | + | + | + | + | + | + | + | + | + | + |
| I2502 | + | + | + | – | – | + | + | + | + | + | + | + | – | |
| I2503 | + | + | + | + | + | + | – | – | – | + | + | + | + | |
| HC26 | I2601 | + | + | + | + | – | + | + | + | + | + | + | + | + |
| I2602 | + | + | + | + | + | + | + | + | + | + | + | + | + | |
| HC27 | I2701 | + | + | – | – | – | + | + | + | + | – | – | + | + |
| I2702 | + | + | + | + | + | + | + | + | + | + | + | + | + | |
| HC28 | I2801 | + | + | + | + | + | + | + | + | + | + | + | + | + |
| I2802 | + | + | + | – | – | + | + | + | + | – | + | + | – | |
| HC29 | I2901 | + | + | + | + | + | + | + | – | – | + | + | + | + |
| I2902 | + | + | + | + | – | + | + | + | + | + | + | + | + | |
| HC30 | I0301 | + | + | – | – | – | + | + | + | + | + | + | + | + |
| I0302 | + | + | + | + | – | + | + | + | + | + | + | + | – | |
| HC31 | I3101 | + | + | + | + | + | – | – | – | – | + | + | + | + |
| I3102 | + | + | + | + | + | + | + | – | – | + | + | + | + | |
| I3103 | + | + | + | – | – | + | + | + | + | – | – | + | + | |
| HC32 | I3201 | + | + | + | + | + | + | + | + | + | + | + | + | + |
| I3202 | + | + | + | + | + | + | – | – | – | + | + | + | + | |
| HC33 | I3301 | + | + | + | + | + | + | + | + | + | + | + | + | + |
| I3302 | + | + | + | – | – | + | + | + | + | + | + | + | – | |
| HC34 | I3401 | + | + | + | + | + | – | – | – | – | + | + | + | + |
| I3402 | + | + | – | – | – | + | + | + | + | + | + | + | + | |
| HC35 | I3501 | + | + | + | + | + | + | + | + | + | – | + | + | + |
| I3502 | + | + | + | + | + | + | + | + | + | + | + | + | + | |
| HC36 | I3601 | + | + | + | + | + | + | + | + | + | + | + | + | + |
| I3602 | + | + | + | – | – | + | + | + | + | + | + | + | + | |
| HC37 | I3701 | + | + | + | + | + | + | + | + | + | + | + | + | + |
| I3702 | + | + | + | + | + | + | – | – | – | + | + | + | + | |
| HC38 | I3801 | + | + | + | + | + | + | + | + | + | – | – | + | + |
| I3802 | + | + | – | – | – | + | + | + | + | + | + | + | – | |
| I3803 | + | + | + | + | – | – | – | – | – | + | + | + | + | |
| HC39 | I3901 | + | + | + | + | + | + | + | + | + | + | + | + | + |
| I3902 | + | + | + | + | + | + | + | – | – | + | + | + | + | |
| HC40 | I0401 | + | + | + | + | – | + | + | + | + | – | – | + | + |
| I0402 | + | + | + | + | – | + | + | + | + | + | + | + | + | |
| HC41 | I4101 | + | + | + | + | + | + | + | + | + | + | + | + | + |
| I4102 | + | + | – | – | – | + | + | + | + | + | + | + | – | |
| HC42 | I4201 | + | + | + | + | + | + | – | – | – | + | + | + | + |
| I4202 | + | + | + | + | + | + | + | + | + | + | + | + | + | |
| HC43 | I4301 | + | + | + | + | + | + | + | + | + | + | + | + | + |
| I4302 | + | + | + | + | – | – | – | – | – | + | + | + | + | |
| HC44 | I4401 | + | + | + | + | – | + | + | + | + | – | – | + | + |
| I4402 | + | + | + | + | + | + | + | – | – | + | + | + | + | |
| HC45 | I4501 | + | + | + | + | + | – | – | – | – | + | + | + | + |
| I4502 | + | + | + | – | – | + | + | + | + | + | + | + | – | |
| I4503 | + | + | + | + | + | + | + | + | + | + | + | + | + | |
| HC46 | I4601 | + | + | + | + | + | + | + | + | + | + | + | + | + |
| I4602 | + | + | + | + | – | – | – | – | – | + | + | + | + | |
| HC47 | I4701 | + | + | + | + | + | + | + | + | + | – | – | + | + |
| I4702 | + | + | – | – | – | + | + | + | + | + | + | + | – | |
| HC48 | I4801 | + | + | + | + | + | + | + | + | + | + | + | + | + |
| I4802 | + | + | + | + | + | + | + | + | + | + | + | + | + | |
| HC49 | I4901 | + | + | + | + | + | – | – | – | – | + | + | + | + |
| I4902 | + | + | + | + | + | + | – | – | – | + | + | + | + | |
| HC50 | I0501 | + | + | – | – | – | + | + | + | + | + | + | + | – |
| I0502 | + | + | + | + | + | + | + | + | + | + | + | + | + | |
| HC51 | I5101 | + | + | + | + | + | + | + | + | + | – | – | + | + |
| I5102 | + | + | + | + | – | – | – | – | – | – | + | + | – | |
| HC52 | I5201 | + | + | + | + | + | + | + | + | + | + | + | + | + |
| I5202 | + | + | + | + | + | + | + | + | + | + | + | + | + | |
| I5203 | + | + | + | + | + | + | + | – | – | + | + | + | + | |
| HC53 | I5301 | + | + | + | + | + | + | + | + | + | – | + | + | + |
| I5302 | + | + | + | – | – | + | + | + | + | – | + | + | – | |
| HC54 | I5401 | + | + | + | + | – | + | + | + | + | + | + | + | + |
| I5402 | + | + | + | + | + | + | + | + | + | + | + | + | + | |
| HC55 | I5501 | + | + | + | + | – | – | – | – | – | + | + | + | + |
| I5502 | + | + | + | + | + | + | + | + | + | + | + | + | + | |
| HC56 | I5601 | + | + | + | + | + | + | + | + | + | + | + | + | + |
| I5602 | + | + | + | + | + | + | + | + | + | – | + | + | + | |
| HC57 | I5701 | + | + | + | + | + | + | + | + | + | – | – | + | + |
| I5702 | + | + | + | + | – | + | + | + | + | + | + | + | + | |
Table 3: Evaluation of Resistance of isolates against common antibiotics using disc diffusion method.
| Isolate No. | Names of Antibiotics | |||||||||||||
| Erythromicin | Tetracycline | Pencillin | Gentamicin | Streptomicin | Amoxicillin | Ciprofloxacin | ||||||||
| Measurement on Zone of Inhibition in (mm) and its Resistance (R) or Sensitivity (S) against Antibiotics | ||||||||||||||
| L. casei | 13 | R | 9 | R | 10 | R | 11 | R | 9 | R | 12 | R | 13 | R |
| L. brevis | 12 | R | 10 | R | 12 | R | 12 | R | 8 | R | 9 | R | 14 | R |
| P. acidilactici | 9 | R | 11 | R | 8 | R | 9 | R | 10 | R | 10 | R | 10 | R |
| L. acetotoleren | 14 | R | 9 | R | 9 | R | 13 | R | 9 | R | 11 | R | 12 | R |
Table 4: Antimicrobial activity of isolated LAB against common pathogens
| Isolate No. | Names of Pathogens | ||||
| E.coli
ATCC-25922 |
P. vulgaris ATCC-33420 | S. aureus ATCC -25922 | S. typhi
ATCC-733 |
P. aeruginosa
ATCC-27853 |
|
| L. casei | 18 mm | 17 mm | 18 mm | 17 mm | 18 mm |
| L. brevis | 21 mm | 14 mm | 16 mm | 19 mm | 20 mm |
| P. acidilactici | 19 mm | 18 mm | 18 mm | 20 mm | 18 mm |
| L. acetotoleren | 20 mm | 16 mm | 15 mm | 16 mm | 16 mm |
4 best species of LAB were screened out of 130 isolates. Identification of LAB was made on the basis of colony morphology, physiological and biochemical tests as per the guidelines mentioned in Bergey’s Manual of Systematic Bacteriology (Hammes et al., 2009). Similar tests performed by earlier researchers found 8 species of LAB that were Gram positive, catalase and oxidase negative and also showed active hydrolysis of arginine (Kang et al., 2019).
The acidic pH of stomach and antimicrobial actions of pepsin provide an effective barrier for LAB to survive in gastrointestinal tract (Kang et al, 2019). For exerting beneficial effects on host, probiotic should be able to maintain its viability along the gastrointestinal transit by surviving under harsh conditions (Tongwa et al., 2019). The survival rate of the isolates of our study were found to be best even at the pH of 2. Traditional techniques of microbiology were used in the study rather than modern molecular techniques because it is more reliable. Modern techniques have some limitations such as the viability of milk microbes cannot be analyzed, total bacteria counts may be over- or underestimated because of cell-wall composition, DNA extraction methods and the number of microbial 16S gene copies which may lead to the over- or underestimation of bacteria counts. Contamination in DNA extraction kit and reagents was also reported in the past studies (Mc Guire, 2015).
CONCLUSION: Human Colostrum contains of large number of bacteria with probiotic potential which greatly helps the infant in boosting up its immunity and in maintaining the gut microbiome. The number of LAB below this count can be a cause of worry for infant. We also found that LAB have the great potentials of fighting against common human pathogens. In our study, we have found that some LAB have great efficiency to resist against antibiotics. Such species of LAB should be commercialized and marketed at a global stage so that problems related to imbalance in gut microbiome can be solved. Through our studies, we also came to know that unnecessary consumption of antibiotics during the time of pregnancy may reduce the LAB count in HC. Therefore, use of antibiotics used be minimized. There are several other facts which are still not known till date such as existence of LAB in HC is still a mystery. LAB in HC is a wide area of research and still needs lots of genuine studies to be carried out to solve the unknown.
ACKNOWLEDGMENTS
Our team is greatly thankful to the entire staff members of Institute for Medical Sciences and Research Center, Jaipur National University (JNU) for directly and indirectly contributing to our studies. A special thanks to the members of Institutional Ethics Committee of JNU for approving our work. We are also thankful to the nursing staff of JNU Hospital especially Mrs. Mamta for helping us in sample collection throughout our entire study.
REFERENCES
Agerholm-Larsen L, Bell ML, Grunwald GK, Astrup A (2002). The effect of a probiotic milk product on plasma cholesterol: a meta-analysis of short term intervention studies, European Journal of Clinical Nutrition. Vol 54 (11), Pages: 856–860.
Ballard O and Morrow AL (2013). Human milk composition: nutrients and bioactive factors. Pediatr. Clin. North Am. Vol 60, Pages: 49–74.
Bishtha, Neha and A.P. Garg (2019). Diversity of Probiotic Bacteria in milk and milk products. In: National Seminar Proceedings on Biodiversity. (In press)
Castellote C, Casillas R, Ramírez-Santana C, Pérez-Cano J, et al., (2011). Premature delivery influences the immunological composition of colostrum and transitional and mature human milk. Journal of Nutrition. Vol 141, Pages: 1181–1187.
Cuello-Garcia C, Fiocchi A, Pawankar R, Yepes-Nuñez J, Morgano GP, et al., (2017). Prebiotics for the prevention of allergies: A systematic review and meta-analysis of randomized controlled trials. Clin. Exp. Allergy (Systematic review). Vol 47 (11), Pages: 1468–1477.
Dunlop AL, Mulle JG, Ferranti EP, Edwards S, Dunn AB, Corwin EJ (2015). Maternal microbiome and pregnancy outcomes that impact infant health: A Review. Adv Neonatal Care. Vol 15(6), Pages: 377-385.
Fern_andez L, Langa S, Martín V, Maldonado A, Jimenez E, Martín R, et al. (2013). The human milk microbiota: origin and potential roles in health and disease. Pharmacol Res. Vol 69 (1), Pages: 1-10.
Guesh Mulaw ,1,2 Tesfaye Sisay Tessema,3 Diriba Muleta ,3 and Anteneh Tesfaye3 (2019). In Vitro Evaluation of Probiotic Properties of Lactic Acid Bacteria Isolated from Some Traditionally Fermented Ethiopian Food Products. Int. J. Micro.(Hindawi). Vol 2019, Pages: 1-11.
Hammes P H and Hertel C, In: Vos P.D., Garrity, G.M., Jones, D, et al. (2009). Bergey’s Manual of Systematic Bacteriology, Springer, New York, Edi 3rd , Pages: 456-479.
Hamilton-Miller JM (2003). The role of probiotics in the treatment and prevention of Helicobacter pylori infection. International Journal of Antimicrobial Agents. Vol 22 (4), Pages: 360–366.
Hunt KM, Foster JA, Forney LJ, Schütte UM, Beck DL, Abdo Z, et al., (2011). Characterization of the diversity and temporal stability of bacterial communities in human milk. PLoS One. Vol 6, Pages: 213-233.
Jin-Sil P, Jeong W, JooYeon J, et a. (2019). Lactobacillus acidophilus Improves Intestinal Inflammation in an Acute Colitis Mouse Model by Regulation of Th17 and Treg Cell Balance and Fibrosis Development. J. Medicinal Food. Vol 21(3), Pages: 1-11.
Jin L, Hinde K, Tao L (2011). Species diversity and relative abundance of lactic acid bacteria in the milk of rhesus monkeys (Macaca mulatta). J Med Primatol. Vol 40(1), Pages: 52-58.
Jost T, Lacroix C, Braegger C, Chassard C (2015). Impact of human milk bacteria and oligosaccharides on neonatal gut microbiota establishment and gut health. Nutr Rev. Vol 73(7), Pages: 426-437.
Kavitha R J and Devasena T (2013). Isolation, Characterization, Determination of probiotic properties of Lactic Acid Bacteria from Human Milk. ISOR J. Phar. and Biolo. Sci. Vol 7(3). Pages: 01-07.
King S, Glanville J, Sanders ME, Fitzgerald A, Varley D (2014). Effectiveness of probiotics on the duration of illness in healthy children and adults who develop common acute respiratory infectious conditions: a systematic review and meta-analysis “. Br. J. Nutr. Vol 112 (1), Pages: 41–54.
Kumari A, Garg PA, Makeen K and Lal M (2008). A bacteriocin production on soya nutri nuggets extract medium by Lactococcus lactis CCSUB202. Int. J. Dairy. Sci. Vol 3(1), Pages: 49-54.
Marchesi JR, Adams DH, Fava F, Hermes GD, Hirschfield GM, Hold G, et al. (2016). The gut microbiota and host health: a Review. Clinical frontier. Gut. Vol 65(2), Pages: 3309-3319.
McGuire MK and McGuire MA (2015). Human milk: mother nature’s prototypical probiotic food? Adv Nutr. Vol 6(1), Pages: 112-123.
Neha B and Garg AP (2019). Antagonistic Activity of Lactic Acid Bacteria Against Common Enteric Pathogens Isolated from Milk and Milk Products and Evaluation of their Probiotic Attributes. Bioscience Biotechnology Research Communications. Vol 12 No. (4) 1173-1184
Pang W and Hartmann P (2007). Initiation of human lactation: secretory differentiation and secretory activation. J. Mammary Gland Biol. Neoplasia. Vol 12, Pages: 211–221.
Pavli F, Tassou C, Nychas E J G and Chorianopoulos N (2018). Probiotic Incorporation in Edible Films and Coatings: Bioactive Solution for Functional Foods. Int J Mol Sci. Vol 19, Pages: 150-168.
Rafter, J (2003). Probiotics and colon cancer. Best Practice and Research Clinical Gastroenterology. Vol 17(5), Pages: 849-859.
Robles-Vera I, Toral M, Romero M, Jiménez R, Sánchez M, Pérez-Vizcaíno F, Duarte J (2017). Antihypertensive Effects of Probiotics. Curr. Hypertens. Rep. (Review). Vol 19 (4), Pages: 26-37.
Shortliffe L and Wein AJ (2013). Infection and Inflammation of the Pediatric Genitourinary Tract. Urology. Saunders Elsevier. Vol 4 (10), Pages: 3121-3143.
Tankoano A, Diop B M, Lingani S H, Mbengue M, Kaboré D, Traoré Y and Savadogo A (2019). Isolation and Characterization of Lactic Acid Bacteria Producing Bacteriocin like Inhibitory Substance (BLIS) from “Gappal”, a Dairy Product from Burkina Faso. Adva. Microbi. Vol 9, Pages: 343-358.
Wang H, Wei XC, Min L and Zhu L (2018). Good and bad: gut bacteria in human health and disease. Reviews. Medi. Biotech. Vol 32 (5), Pages: 1075-1080.