Department of Biological Science, Faculty of Science, King Abdulaziz University Jeddah, Saudi Arabia.
Corresponding author email: balaidaros@kau.edu.sa
Article Publishing History
Received: 18/09/2021
Accepted After Revision: 15/12/2021
In the past two decades, phenolic compounds have had different applications, however their use in densification has increased considerably due to Covid 19. Discharge of these dangerous materials is highly toxic and causes risk and severe problems to the environment and health of human and animals, in addition to it being harmful to the aquatic life. Phenol degradation is very important due to high toxicity and stability. The aim of this study is to isolate phenol-degrading aerobic bacteria from hydrocarbon contaminated soil or wastewater, collected from the industrial area of Jeddah. Minimal medium containing phenol as carbon source was used to isolate different bacteria. About 30 actinomycete isolates were obtained, purified and preserved on Starch nitrate.
Out of 30 isolates, eight isolates (27%) grow well in medium containing 0.1% phenol. After growing in broth medium, isolate BA4 and isolate BA8 were very active in phenol degradation. Growth and phenol degradation was measured in liquid medium for the two isolates. Morphological and physiological characters of these isolates were detected using different methods. Using molecular methods, they were belonging to a genus of actinomycetes. They were identified as Streptomyces flavabus BA4 and Streptomyces sp. BA8.The effects of some growth factors on growth and phenol degradation were determined. Growth was measured by dry weight (mg/l) while phenol degradation was detected by assaying the residual phenol concentration.
The presence of electron donors such as glucose, starch, glycine, peptone, and Na acetate affect both growth and phenol degradation. It was clear that addition of 1 g/l peptone enhanced both growth and phenol degradation. The isolate use phenol and its derivatives m-cresol and o-cresol as carbon sources and addition of vitamin B complex increased the bacterial growth. In conclusion, phenol degradation was detected by actinobacteria and was affected by some physical and biochemical factors. It was noticed that optimization of growth conditions enhanced both growth and phenol degradation by the two selected Streptomyces isolate. Degradation process by isolate BA4 could be a promising solution for removal of phenol from wastewater.
Phenol, Streptomyces, Degradation, 16srrna, Wastewater
Alaidaroos B.A. Isolation and Molecular Identification of Phenol Degrading Bacterium from Industrial Wastes, Collected from Jeddah Saudi Arabia. Biosc.Biotech.Res.Comm. 2021;14(4).
Alaidaroos B.A. Isolation and Molecular Identification of Phenol Degrading Bacterium from Industrial Wastes, Collected from Jeddah Saudi Arabia. Biosc.Biotech.Res.Comm. 2021;14(4). Available from: <a href=”https://bit.ly/3FfG58K“>https://bit.ly/3FfG58K</a>
Copyright © Alaidaroos This is an open access article distributed under the terms of the Creative Commons Attribution License (CC-BY) https://creativecommns.org/licenses/by/4.0/, which permits unrestricted use distribution and reproduction in any medium, provide the original author and source are credited.
INTRODUCTION
Increasing population and industrialization has critical effects on human beings and the environment. Many studies have proved that these industrial effluents are the main source of many kinds of pollution of natural water. In the effluents of major industries, phenolic materials are present as dangerous pollutants especially in oil refineries, petrochemical plants and industrial effluents of paper mills. Paper industries produce a huge amount contaminated water with organic and inorganic pollutants in addition to coloring materials which destroy soil and growing plants in these soils (IARC, 1989, Mörsen and Rehm, 1990, Baruah et al. 1996, Gerginova et al., 2007, Hussain et al., 2008, 2009, Hussain et al., 2010, Alhazmi et al., 2018, Xu et al., 2021).
Several studies have been conducted to examine the effluent constituents of textile, dyes, coal processing and plastics and pharmaceutical industries in addition to the effluents of pulp and paper, oil refineries, polymeric resins, insecticides, pesticides and steel plants. They have reported that these effluents contained phenolic compounds and phenol is the main pollutant which destroys the skin, cause vomiting, paralysis, lung failure and cardiac arrest, modify the water taste and odor.LD50 of these dangerous materials to fishes and human beings have been found to be 5–25 and 10–24 mg/l, respectively being potentially carcinogenic at these concentrations, (Pazarlioglu and Telefoncu, 2005, Shazryenna et al 2015, Xu et al., 2021).
Due to the high toxicity of phenol and its derivatives, several regulatory laws all over the world such as MOEF, GOI, EPA and USEPA have reported them as the main dangerous pollutants with low permission limits in water and before discharge contaminated waste water into any natural sources, removal of phenol is prominent and urgent for developing green and sustainable environments.Many chemical methods like adsorption, extraction, ion exchange, oxidation, polymerization and coagulation are applied for removal of phenol up to the limit of WHO recommendation for drinking water but these methods were not efficient or effective. Biological methods are the solution for wastewater treatment and reducing the poisonous organic compounds (Sonawane and Koreke 2016, Mallick et al., 2021).
Several microorganisms can be used to treat phenol contaminated effluents by mainly techniques including phenol degradation enzymes like peroxidase and laccase and biosorption on live or dead cells. Untreated effluents used to be pumped to the Red sea cause a negative impacts on marine environments. Studies must be planned for isolation and screening of phenol-degrading bacteria from contaminated water effluent to decrease pollution (Park et al., 2007, Malaviya and Rathore, 2007, Dubey and Hussain 2014, Xu et al., 2021).
Saudi Arabia is a big country, located in the arid area and its area is about 2.15 million km2. Its problem is water, where about 80% of the used water is from shallow grounds which can be contaminated by the phenolic materials or heavy metals. Therefore, it is expected that the situation of groundwater contamination may become more alarming in the coming years, mostly because of the rapid industrialization and urbanization in this country, (Al-sefry and Sen, 2006, Mallick et al., 2021).
In the effluents of different petrochemical, pharmaceutical and paint industries, phenolic products are found which are classified as highly hazardous chemicals that cause dangerous problems to man, animals and aquatic and terrestrial environments. In animals, gastrointestinal irritation, diarrhea, weight loss and liver and kidney toxicity have been naoted after exposure to phenol. Contamination with the priority pollutant, phenol in waste water must be limited for good natural system-functions and sustainable environment. All International regulatory bodies have reported that phenol level must be brought to the required levels not exceeding 9–25 mg/l, which is the toxic level for living cells (APHA, 2005, Mallick et al., 2021).
It was also observed that chemical contamination is limited to shallow aquifers only while the deeper aquifers are safe. Besides, it is important to emphasize the presence of phenolic compounds in groundwater.
Therefore, there is a need to carry out a systematic and exhaustive study of the toxic materials in the groundwater of the country. The efforts are made to describe the groundwater in KSA, toxicities of the metal ions in the groundwater, sources of metal ions contamination and the future challenges and the remediation measures needed to protect groundwater resources. Certainly, studies about phenol degradation will be important for environmental and the regulatory authorities. Bacteria, fungi and actinomycetes have the ability of using phenol as carbon source and metabolized it to CO2 and water. Phenol-degrading microorganisms which successfully completely removed phenol from waste water are still needed to be discovered (Xu et al., 2021).
Considering the above-mentioned facts, this study was aimed to carry out the isolation and molecular identification of promising potential actinobacteria from contaminated soil for phenol degradation and optimization the degradation conditions for maximum activities in treating phenolic wastewater.
MATERIAL AND METHODS
Contaminated soil (10) and wastewater (10) samples were collected from Wastewater treatment plant, Jeddah industrial city, Jeddah, Saudi Arabia (Figure 1). They were collected either in sterile plastic bags or sterile bottles. Soil samples were air dried and sieved. The bacteria from contaminated soil or wastewater samples were obtained on starch-nitrate agar medium which was adjusted to pH 7.0 and contained (g/l): starch, 20; KNO3, 2; K2HPO4, 1; KH2PO4, 0.7; MgSO4·7H2O, 0.7; agar, 20 after 7 days of incubation at 37◦C (). All the obtained bacterial isolates were purified and preserved on the same medium on slants at 4◦C until used (Arifuzzaman et al., 2010, Tork et al., 2018, Aly and Tork, 2018, Khalel et al., 2021).
Figure 1: The collection sites of the samples obtained
from industrial area, Jeddah.
Different bacterial isolates (30) were screened on mineral salt agar medium containing Phenol as carbon source (100 mg/l) for 7 days at 37oC and the most active isolates that showed the highest growth (8 isolates) were selected and screened in liquid broth medium and Phenol degradation was determined (Ali et al. 1998). The eight bacterial isolates that showed growth in the presence of phenol were screened in 250 ml Erlenmeyer flasks containing 50 ml of the basal Mineral salt medium broth medium supplemented with phenol (0.1 g/l) as a sole carbon source for 7 days.
The medium pH was adjusted to pH 7. Each flask was inoculated with 2 ml of fresh prepared bacterial suspension, containing 6 x 106 cfu/ml and incubated in shaking incubator at 37oC and 120 rpm. At the end of the growth period, the growth was determined as g/l. Cells were harvested by centrifugation at 5,000 rpm for 5 min, washed dried at 60◦C for 3 days and weighted. Phenol degradation was measured quantitatively in the culture filtrate by the increase in the absorbance using spectrophotometric method. All experiments were made in triplicate and averages were calculated.
In any contaminated area, the amount of phenol was detected using 4-aminoantipyrine reagent and UV–Vis spectrophotometer (Systronics UV–Vis spectrophotometer 118) as described by APHA (2005), Sachan et al. (2019). All the chemicals used in present study were obtained from Hi- Media Laboratories Pvt. Ltd, Mumbai, India
The isolates BA4 and BA8 were grown on MSA medium containing different concentrations of Phenol (100 -1300 mg/ml). The plate was inoculated with 1 ml of the bacterial suspension (6×106 CFU/ml ), previously grown for one week in starch nitrate broth medium at 120 rpm for 4 days. The inoculum was spread over the surface of the medium. Incubation was then carried out at 37°C. Assessment of degradation activity was detected by measuring the degree of growth, heavy, moderate or poor.
The two selected bacterial isolates were characterized and identified. The selected isolates were identified according to International Streptomyces Project (Shirling and Gottlieb 1966). The growth, color of the aerial mycelia was determined on different agar media. Also, the growth of the two isolate on different carbon and nitrogen sources were determined (John et al., 2000, Abd-El-Haleem et al., 2002). DNA was extracted and 16SrDNA genes of the active isolates were purified and sequenced The sequence was analyzed and compared to 16SrDNA genes in the GenBank databases (Weisberg et al.,1991, Xu et al., 2021).
Staphylococcus aureus, Bacillus cereus, E. coli (MTCC 443), and Pseudomonas aeruginosa (MTCC 8076) were used for antimicrobial study. All the stock cultures were collected from KAU hospital. All of the bacterial strains were grown and maintained on their specific medium. The bacteria were sub-cultured overnight for further use. The antimicrobial activity of the isolate BA4 and AB8 was determined by agar disk diffusion method. Cotton swab was used to inoculate the surface of nutrient agar plate and the plate was allowed to dry. Using a sterilized forceps, a disk (5 mm) of the bacterial growth was transferred onto the agar surface (Agwa et al., 2000). Amoxicillin was used as control. The experiment was conducted in triplicates. The plates were incubated at 37°C for 24 hrs. At the end of the period, the inhibition zone against agar disc was measured (mm).
For maximum Phenol degradation, growth conditions were optimized. The degradation of Phenol was performed using 50 ml of Mineral salt medium (pH 7.0) with 0.1 % Phenol as a carbon source in250 ml Erlenmeyer flasks. All flasks were incubated at 120 rpm for 7 days.The effect of different electron donors such as glucose, glycine, peptone, and Sodium acetate(1%) was recorded and phenol reduction was estimated following usual method. Effect of different concentrations of peptone (0.5, 1.0, 1.5, 2.0 and 2.5 g/l)on growth and Phenol degradation were determined.
In the presence of phenol (0.1 g/l), effect of some additives like vitamin B1, B6, B12, mixture of B1 + B6 + B12, indole acetic acid, nicotinic and tryptophane at concentration of 0.1 g/100 ml on growth of isolates BA4 and BA8 was determined in starch nitrate medium after7 days of incubation at
The statistical Package for Social Science (SPSS for windows, version 16) (SPSS Inc., Chicago, IL, USA) was used for data analyses and all results were expressed as mean ± SD .Triplicate measurements were carried out in all the cases and t-test was used to detect any significant difference between sample and control. Significant results were obtained at P<0.05.
RESULTS AND DISCUSSION
From the contaminated area in the industrial zone in Jeddah, Saudi Arabia, twenty contaminated soil and Wastewater samples were collected. Using dilution plate method, 30 different bacterial isolates were obtained on Starch Nitrate Agar. All the isolates were purified and preserved on the previous medium. The colonies of the isolates had different color and shapes and all isolates had filamentous shape and showed Gram positive reaction. On MSA medium containing 0.1% phenol, all isolates were screened for phenol degradation.
The degree of growth was determined and out of thirty isolates, eight isolates (27%) showed good, moderate or poor growth on the solid agar medium with phenol as carbon source (Table 1). Also, all the eight isolates were Melanin pigment producer while 4 isolates were Diffusible pigment producers. Moreover, the eight active isolates were grown in liquid medium and growth and residual phenol were determined. In liquid medium, all the tested isolates use phenol as carbon source with different degree (Table 2).
The best degradation was noticed by the lowest absorbance for phenol assay. In liquid medium, the isolates BA47and BA8showed the best dry weight per liter using phenol as carbon source and decreased the phenol concentration in the medium. The two isolates were selected for more detail studies. Morphological, physiological and biochemical characterization using different methods were carried out. The growth on starch nitrate agar of the two isolates and the shape under light and electron microscope were reported in Figure 2.
Isolate BA4 had pink color while the isolate BA8 had white color. The two isolates had aerial and substrate mycelia and spore chain. The spore chain of isolate BA4 was long and the numbers of spores was ranged from 20 to 30 per chain. The spore was spherical with spiny surface. The spore chain of isolate BA8 was long and the numbers of spores was ranged from 30 to 40 per chain. The spore was cylindrical with smooth surface. Table 3 showed the morphological characteristics of the two selected isolates. Growth and color of the aerial mycelia of isolate BA4 and isolate BA8 on different growth media after growth for 10 days at 30℃ were summarized in Table 4.
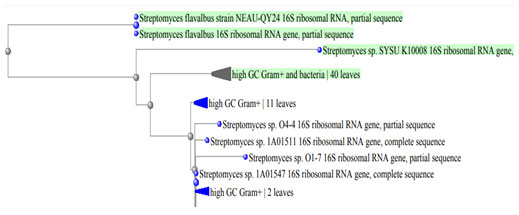
Moreover, Table 5 showed the growth of the two bacterial isolates BA4 and BA8 on different carbon and nitrogen sources. Also, the antimicrobial activities (diameter of the inhibition zone, mm) of the two tested bacterial isolates against different bacterial pathogens and compared o control were shown in Table 6. From the previous results, the two isolates were belong to genus Streptomyces. Identification of the two tolerant isolates was confirmed using molecular methods and they were belonging to the same genus which was belonging to actinomycete genera. They were identified as Streptomyces flavabus BA4 and Streptomyces sp. BA8. Phylogenetic tree based on 16s rRNA of the two selected Streptomyces isolates was detected (Figure 3).
Table 1. The isolated Gram positive bacteria, color of aerial and
substrate mycelia, growth and diffusible pigment production
</tbody border=”1″>
| Bacterial isolate | Source of isolation | Colony
color |
Growth | Diffusible pigment | Melanin pigment |
| BA1 | Wastewater | White | ++ | -ve | +ve |
| BA 4 | Wastewater | Pink | ++++ | +ve | +ve |
| BA 6 | Wastewater | Gray | ++ | -ve | +ve |
| BA 8 | Wastewater | Gray | ++++ | +ve | +ve |
| BA 15 | Wastewater | Gray | ++ | -ve | +ve |
| BA 17 | Soil | Gray | ++ | -ve | +ve |
| BA 19 | Soil | Gray | ++ | +ve | +ve |
| BA25 | Soil | Gray | ++ | +ve | +ve |
| ++++: high growth, ++: moderate growth, +: poor growth, +ve: pigment present ,
-ve: pigment absent |
|||||
Table 2. Growth and phenol degradation by the selected actinomycete isolates
| Phenol concentration
Bacterial isolate |
500 mg/l | 1000 mg/l | ||
| Growth
(Dry weight, mg/ l) |
A235 | Growth
(Dry weight, mg/ l) |
A235 | |
| BA1 | 0.21±3.04 | 0.10 ±0.06 | 0.ND | ND |
| BA 4 | 0.89 ±5.06 | 0.16 ±0.08 | 0.48 ±5.06 | 0.16 ±0.08 |
| BA 6 | 0.19 ±6.07 | 0.11 ±0.07 | 0. ND | ND |
| BA 8 | 0.82 ±6.07 | 0.21 ±0.9 | 0.32 ±6.07 | 0.11 ±0.07 |
| BA 15 | 0. 29±2.09 | 0.10 ±0.01 | ND | ND |
| BA 17 | 0. 25±2.09 | 0.15 ±0.04 | ND | ND |
| BA 19 | 0. 33±8.02 | 0.14 ±0.08 | ND | ND |
| BA25 | 0. 39±3.07 | 0.14 ±0.03 | ND | ND |
Figure 2 A: The isolate BA4 and B: the isolate BA8, a: on starch nitrate agar,
b: under electron microscope, c: under light microscope.
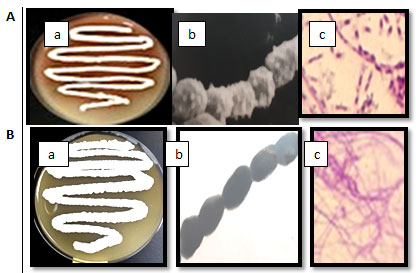
Table 3. Morphological characteristics of the two selected isolates
| Characters | Isolate BA4 | Isolate BA8 |
| Source | Waste water | Waste water |
| Gram stain | Gram-positive | Gram-positive |
| Color | Pink | Yellow |
| Motility | Absent | Absent |
| Respiration | Aerobic | Aerobic |
| Substrate Mycelium | Branched | Branched |
| Spore chain | Positive | Positive |
| Motile spores | Absent | Absent |
| Aerial and substrate mycelia | Present | Present |
| Optimum
temperature |
30 °C | 37 °C |
| Optimum pH range | 6.5 – 7.0 | 6.5 – 7.5 |
| Catalase | Positive | Positive |
| Penicillin | Sensitive | Sensitive |
| Cephalosporin | Resistance | Resistance |
Table 4. Growth of isolate BA4 and isolate BA8 on different growth media for 10 days at 30℃.
|
Media |
Isolate BA4 | Isolate BA8 | ||
| Growth | Color of aerial mycelia | Growth | Color of aerial mycelium | |
| Starch Nitrate agar | Heavy | Pale Pink | Heavy | Light yellow |
| Yeast extract-malt extract agar (ISP-2) | Moderate | Pink | Heavy | Light brown |
| In-organic salt-starch iron agar (ISP4) | Heavy | Dark brown | Heavy | Creamy |
| Glycerol asparagine agar (ISP-5) | Moderate | White | Moderate | Yellow |
| Tyrosine agar (ISP-7) | Heavy | Light brown | Heavy | Pale yellow |
| E-Medium (ISP-9) | Moderate | Yellow | Moderate | Creamy |
Table 5. Growth of bacterial isolates BA4 and BA8 on different carbon and nitrogen sources
| Carbon source | isolateBA4 | Isolate
BA8 |
Nitrogen source | isolateBA4 | Isolate
BA8 |
| Glucose | +++ | +++ | Ammonium sulfate | ± | ± |
| Sucrose | + | + | Ammonium chloride | + | + |
| Starch | +++ | ++ | Potassium nitrate | + | + |
| Lactose | + | + | Glycine | + + | + + |
| Vanillin | ± | ± | Dextrose | + | + |
| Peptone | +++ | +++ | Maltose | + | + |
+++: high utilization, ++: moderate utilization, +: weak utilization, ±: very weak utilization
Table 6 .The antimicrobial activities (diameter of the inhibition zone, mm) of the two tested
bacterial isolates against different bacterial pathogens and compared o control.
| Tested pathogen | Isolate BA4 | Isolate BA8 | Amoxicillin |
| Staphylococcus aureus | 19±3.1 | 17±3.1 | 21±4.11 |
| Bacillus cereus | 23±3.1 | 20±3.1 | 29±2.10 |
| E. coli (MTCC 443) | 23±3.1 | 20±3.1 | 29±3.19 |
| Pseudomonas aeruginosa | 17±3.1 | 16±3.1 | 21±3.11 |
Figure 3: Phylogenetic tree based on 16s rRNA of the two selected Streptomyces isolates.
Tolerance of the two isolates to phenol was recorded on starch nitrate agar containing increasing concentration of phenol. It was found that increasing phenol concentration decreased the growth up to 1200 mg/l where there is no growth (Table 7). The growth of the selected isolate BA4 with increased concentrations of phenol (0.1- 1.2%) was shown in Figure 4. The isolate BA4 was grown in MS broth medium with phenol as carbon source. It was noted that at low phenol concentration the growth was maximum and increasing phenol decreased the growth. No growth was recorded at 1300 mg/ml phenol. The quantity of phenol present in the solution increased with increasing the used phenol concentration (Figure 5).
The effects of some growth factors on growth and phenol degradation were determined. Growth was measured by dry weight (mg/l) while phenol degradation was detected by assaying the residual phenol concentration. The addition of some electron donors such as glucose, starch, glycine, peptone, and Na acetate on growth and Phenol degradation was determined. It was clear that addition of these materials significantly enhanced both growth and phenol degradation except starch which showed no significant differences compared to control (Figure 6). The best results were obtained with peptone. It was clear that addition of 1 g/l peptone enhanced both growth and phenol degradation. Increasing peptone concentration decreased phenol degradation by the tested bacterium BA4 (Figure 7). The isolate BA4 can use phenol and its derivatives m-cresol and o-cresol as carbon sources and addition of vitamin B complex increased the bacterial growth and phenol degradation (Table 8).
Table 7. Tolerance of the two selected isolates to different Phenol concentrations.
| Phenol concentration (mg/l) | 100 | 200 | 400 | 800 | 1000 | 1200 | 1300 |
| Isolate BA4 | +++ | +++ | +++ | ++ | ++ | + | – |
| Isolate BA8 | +++ | +++ | ++ | ++ | ++ | – | – |
+++: high utilization, ++: moderate utilization, +: weak utilization, -: no utilization
Figure 4. The growth of the selected isolate BA4 grow on starch nitrate
medium with increased concentrations of phenol (0.1- 1.2%).
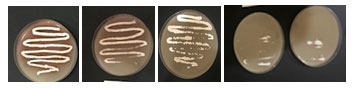
Figure 5. Growth and Phenol degradation by the isolate BA4
grownin different concentration of phenol
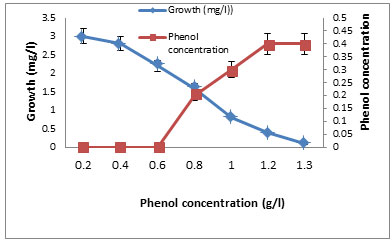
Figure 6. Growth andPhenol degradation by the isolate BA4 grownin
medium subdifferent concentration of phenol
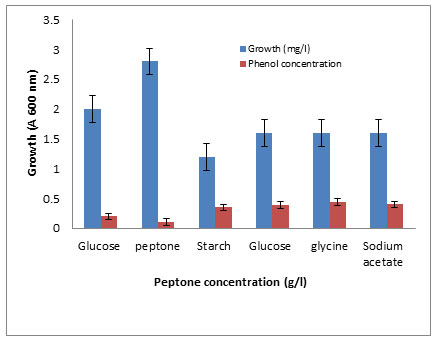
Figure 7. Growth and Phenol degradation by the Actinomycete isolate BA4
grown in different concentrations of peptone
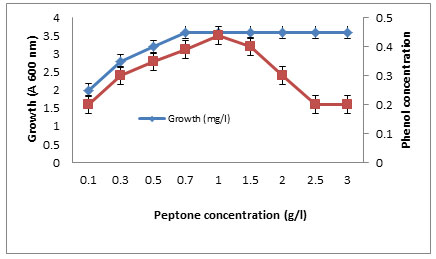
Table 8. Effect of some activators on growth of BA4 on starch nitrate
medium containing phenol or its derivatives.
| Tested material (0.1 g/100 ml) | Diameter of colony (mm) | ||
| Phenol | m-cresol | o-cresol | |
| Control | 30.0±5.0 | 17.9±5.3 | 17.0±5.0 |
| B1 | 37.9±3.1* | 17.4±5.3 | 17.9±5.0 |
| B6 | 33.9±2.5 | 17.5±5.1 | 17.6±5.0 |
| B12 | 33.3±5.0 | 23.2±5.0 | 16.9±3.0 |
| B1+B6+B12 | 39.3±5.0* | 29.2±5.0* | 19.9±1.3 |
| Indole acetic acid | 30.5±1.3 | 17.3±3.1 | 12.4±2.0* |
| nicotinic acid | 27.0.9±1.4 | 17.0±2.0 | 17.5±1.5 |
*: significance P≤5%
The currently methods for removing and degradation of toxic wastes and chemicals that harm human and animal health by bacteria or fungi are effectively used. Industrial effluents are mainly contained phenol and/ or phenolic compounds that must be safely bio- removed or biodegraded. In wastewater, presence phenol cause severe problems during treatment process, thus phenol biodegradation is a necessary process in the wastewater treatment process. Wastewater is unique environment due to the extreme conditions that is preventing easy growth. In the contaminated area, phenol bioremediation and studying the intermediate compounds are needed (Borghei and Hosseini, 2004; Abd El-Zaher et al., 2011, Xu et al., 2021).
Nakagawa et al. (1963) isolated and characterized catechol oxygenase from Brevibacterium fuscum for biodegradation of phenol. Isolation of bacteria for biodegradation of phenolic compounds from wastewater or polluted soil has been reported by Tallur et al. ( 2006) and Atsushi et al. (2006). Khleifat and Kaled, (2007) isolated Actinobacillus species that degraded phenol. Similarly, Nagamani et al. (2009) isolated and identified Xanthobacter flavus for removal of phenolic materials while Nair (2007, 2008) tried to purify paper factory effluent using a phenol degrading Alcaligenes sp. Also, Nilotpala and Ingle (2007) mineralized phenol by Serratia plymuthica strain GC isolated from sludge sample while Hasan and Jabeenb (2015) isolated Pseudomonas sp. and Bacillus subtilis from malathion and phenol contaminated soil
In this study, waste water and soil were used as source of phenol degraded bacteria. Similarly, Mohite et al. (2010) used contaminated soil sample for isolation of Streptococcus epidermis which use was isolated on medium containing phenol as carbon and energy source.
Recently Sachan et al., (2019) havex reported removal of phenol from waste-water by two bacterial isolates up to 1800 mg/L. Starch nitrate agar was used for growing and maintained the two selected isolates. The previous medium was used by many authors to isolate and grow Actinobacteria (El-Naggar et al., 2014, Aly et al., 2018). On MSA medium containing 0.1% phenol, eight isolates (27%) showed growth and all the eight isolates were melanin pigment producers. Melanin pigments are natural biopolymers which have special biological activities and protect organisms from difficult environmental conditions. During the past decade, melanins have attracted increasing attention for their use in drug delivery, photoprotection and environmental bioremediation (Tran-Ly et al., 2020).
Using enrichment technique, two bacterial isolates BA4 andBA8 were selected for their maximum ability to degrade phenol. Similarly, isolate ABO11 used phenol (0.8 g/l) as carbon and energy source (Aburas, 2016) while lower degradation was obtained by Mohite et al. (2010), who recorded degradation of 200 mg/l phenol. Phenol removal was generally by oxidation using phenol hydroxylase to catechol.
The two active bacterial isolates BA4 and BA8 were belonging to Gram-positive Actinobacteria which was worldwide in a variety of natural habitats. Actinobacteria are rich with a high guanine plus cytosine (55-70 %) content and they were extremely diverse group of microorganisms. Their members differed in the chemical structure of the cell wall, cell morphology, and physiological characters (Singh et al., 2016, Tran-Ly et al., 2020).
In this study, the two isolates had hyphae that branch, generating aerial mycelium with lengthy chains of spores. They were identified according to phenotype and genotype characterization. Sequencing of the 16S rDNA gene shared 93 and 95% identity with that of genus Streptomyces, respectively. They were identified as Streptomyces flavabus BA4and Streptomyces sp. BA8. Phenol treatment was applied for isolation of rare Actinomycetes from soil samples which was suspended in 1.5 % (w/v) phenol solution at 30◦C for 30 minutes they boated 61 isolates, and most of them were not Streptomyces (only 24.6%), whereas other genera such as Micromonospora, Actinomadura, Microbispora and Polymorphospora were isolated with ratios of 49.2%, 13.1%, 9.8%, and 3.3%, respectively (Istianto et al., 2012). The results of Azadi and Shojaei (2020) showed that a range of Nocardia species that belonging to actinobacteria have not received much attention but have great potential for bioremediation purposes.
Phenol biodegradation with least two bacterial enzymes, involved in the process, is widely used through definite methods. Also, Candida tropicalis was used by Mohd and Piakong (2006) for phenol biodegradation while Shawabkeh et al., (2007) reported that within 72 hr Klebsiella oxytoca degraded 75% of 100 ppm of phenol. Adjusting growth conditions improved phenol degradation process. In this study, addition of 1% peptone enhanced both growth and phenol degradation using Succinic acid and glycine as carbon and nitrogen source. Capacity of Pseudomonas aeruginosa for phenol biodegradation was enhanced of addition of different organic nitrogen sources (Aspartic acid, Beef extract, Peptone, Tryptone and Yeast extract.
They added that maximum phenol degradation was obtained using yeast extract and peptone addition to the used medium while moderate degradation was obtained with the other nitrogen sources. Wastewater had unpleasant smell and brown color due to the presence of high concentration of phenol and its derivatives (Madan et al. 2018) which have direct inhibitory effects on some bacteria. These materials are not easily biodegraded and cause increase the COD, suspended and dissolved and total dissolved solids in such type of wastewaters. The discharge of waste water into water bodies may cause a drop or increase in their pH values due to the size and activities of microbial population.
High biochemical oxygen demand and chemical oxygen demand indicated the presence of high concentration of organic matter, thus treatment of wastewater is required before discharging. Biological treatment using useful microfora especially bacteria is the only choice for treatment (Marihal and Jagadeesh 2013). Phenol-degrading enzymes are broadly distributed in different microorganisms that play an important role in the degradation of phenol. (Marihal and Jagadeesh, 2013, Haritash and Kaushik, 2009). Bacterial isolation by enrichment method was used to obtain specific bacteria that degrade phenol (Sachan et al., 2019).
They added that many phenol tolerant bacteria and fungi were isolated from phenol contaminated area and they identified 16 bacterial isolates from paper effluent sample that were grow on MSM broth medium with phenol. At higher concentration of phenol (2000 mg/l), negligible growth was recorded which was due to bacteria sensitivity to higher concentration of phenol. Some isolates required acclimatization on different concentrations of phenol (Abd-El-Haleem et al., 2002).
Gradually, bacteria adapted themselves to degrade wastes (Arutchelvan et al., 2005, Sun et al., 2012). After acclimatization, bacteria showed surprising ability in fast reproduction and phenol removal. Annadurai et al., (2000a, b) reported that absorption and biodegradation of phenol was increased by using Chitosan immobilized Pseudomonas putida. Screening by growth studies on medium with phenol lead to isolation of species with high capability of phenol removal that can be utilized to purify wastewaters, containing high phenol concentrations.
Similar to our results, Yang and Lee (2007) reported that phenol has a potentially inhibitory effect on cell growth. Two bacterial isolates SP-4 and SP-8 showed good growth minimal salt medium with phenol in the presence of 1% glucose (w/v), whereas no growth has been observed in the absence of glucose. Both the strains showed fast and luxuriant growth at phenol concentration of 0–1000 mg/l. Isolate SP-4 is tolerate phenol up to 1600 mg/L while isolate SP-8 can tolerate the phenol up to 1800 mg/l and no growth has been observed for the two bacterial isolates at 2000 mg/l phenol.
Information about degradation potentials of bacteria, isolated from polluted places, is essential in designing stable bioremediation methods (Azadi and Shojaei, 2020). Majority of studies has deals with isolation of actinomycetes from normal habitats and determine their biological activities while actinomycetes that live in unusual environments was not studied well and are unexplored (Singh et al., 2016, Aly et al., 2020, Bahamdain et al., 2020).. Isolation of actinobacteria for cleanup soil of pesticides, metals, and mixed pollution had been reported. Due to their high catabolic ability and durability in harsh conditions and polluted area, screening new regions for unexplored actinomycetes discovered novel isolates with excellent and significant bioremediation abilities (Azadi and Shojaei, 2020).
The xenobiotic material p-Nitrophenol is dangerous and highly toxic to soil microflora, released into soil after pesticides degradation organophosphate. Two actinomycete isolates A1 and A5 were found to be promising PNP bio- degraders. Isolate A5 was identified as Streptomyces coeruleorubidus and optimization growth conditions can improve the biodegrading abilities of the selected actinomycete isolate (Jaweria et al., 2013). For removal of toxic chemical and pesticides from contaminated soil and wastewater effective system is needed. Inorganic compounds are difficult to be removed while organic compounds were completely degraded to less toxic materials. Using some bacterial genera in bioremediation process are used to solve these problems.
Actinobacteria are excellent choice due to their high presence in soil and water and they previously maintained ecological balance between soil flora. Several species of Actinobacteria have been found to use phenol and pesticides as carbon sources, completely degrading them to nontoxic compounds (Fuentes et al., 2010). For example, several strains of Streptomyces (including Streptomyces espinosus) have been found to produce tyrosinase enzymes, which are helpful and more effective than that obtained from other bacteria in the removal of phenols, a component of many pesticides that polluted water and soils. In addition, these bacteria produced secondary products that can be used for some pollutants removal, thus they are excellent candidates for organic pollutants removal without damage the environment (Myronovskyi et al., 2020).Finally, we can conclude that some Actinobacteria, isolated from contaminated region can be used in cleaning the environment and waste water from phenol pollution.
CONCLUSION
Phenol degradation was detected by actinobacteria and was affected by some physical and biochemical factors. It was noticed that optimization of growth conditions enhanced both growth and phenol degradation by the two selected Streptomyces isolate. Degradation process by isolate BA4 could be a promising solution for removal of phenol from wastewater. Its potential for use in biological treatment of phenol in industrial wastewater must be studied to improving the quality of the final wastewater. These results provided useful information about use of the phenol-degrading bacteria for cleaning of industrial wastewater and the quality of the resulting wastewater will be improved. In occlusion, the degradation of phenol helps to overcome some of the pollution problems associated with the use of detergent.
ACKNOWLEDGEMENTS
This Project was funded by the Deanship of Scientific Research (DSR) at King Abdulaziz University, Jeddah, Saudi Arabia, (Grant number J 32- 247-1439). The author therefore acknowledges DSR for technical and the financial support.
Conflict of Interests: The author declares no conflict of interest exist.
REFERENCES
Abd El-Zaher EH, Mahmoud YA and Aly M M (2011). Effect of different concentrations of phenol on growth of some fungi isolated from contaminated soil. African Journal of Biotechnology, Vol. 10(8), pp. 1384-1392.
Abd-El-Haleem D, Moawad H, Zaki EA, Zaki S (2002). Molecular characterization of phenol-degrading bacteria isolated from different Egyptian ecosystems. Micro Ecol., 43:217–224
Aburas MA (2016).Removal of Phenol Using Spore Forming Bacillus ABO11 Isolated from Waste Water Treatment Plant. Advances in Microbiology, Vol.6 No.12: 123-139.
Agwa A, Aly MM, Bonaly R (2000). Isolation and characterization of two Streptomyces species produced non polyenic antifungal agents. J. Union Arab Biol., Vol.7 B, pp.: 62- 84.
Alhazmi NM, Ghanem KM, El Ahwany A, Aly M and Bokhari FM ( 2018). Degradation of Poly-β-hydroxybutyrate by Trichoderma asperellum. Journal of Experimental Biology and Agricultural Sciences, Vol. 6(2): 324 – 334.
Ali S, Lafuente RF, Cowan DA (1998). Meta-pathway degradation of phenolics by Thermophilic Bacilli. Enz Microb Technol., 23:462–468
Al-sefry SA. and Sen Z (2006). Groundwater rise problem and risk evaluation in major cities of arid lands- Jedddah Case in Kingdom of Saudi Arabia. Water Resources Management, 20: 91-108.
Aly MM and Tork S (2018). High keratinase production and keratin degradation by a mutant strain KR II, derived from Streptomyces radiopugnans KR 12. Journal of Applied Biological Sciences, 12 (1): 18-25.
Annadurai G, Balan MS, Murugesan T (2000a). Design of experiments in the biodegradation of phenol using immobilized Pseudomonas pictorium (NICM – 2077) on activated carbon. Bioproc. Eng. 22: 101-107.
Annadurai G, Rajesh S, Mahesh KP, Murugesan T (2000b). Absorption and biodegradation of phenol by Chitosan immobilized Pseudomonas putida (Mcm.2174). Bioproc. Eng., 22: 493-501.
APHA (2005). Standard methods for the examination of water and wastewater, (20th ed), Washington DC, USA.
Arifuzzaman M, Khatun MR and Rahman H (2010). Isolation and screening of actinomycetes sundarbans soil for antibacterial activity. Afr. J. Biotechnol., V9, P: 4615-4619.
Arutchelvan V, Kanakasabai S, Nagarajan S, Muralikrishnan V (2005). Isolation and identifcation of novel high strength phenol degrading bacterial strains from phenol formaldehyde resin manufacturing industrial wastewater. J Hazard Mater 127:238–243.
Atsushi S, Yasushi I, Arata K (2006). Aerobic and anaerobic bio degradation of phenol derivatives in various paddy soils. Sci. total Environ., 367: 979-987.
Baruah NK, Kotoky P, Bhattacharyya KG and Borah GC (1996). Metal speciation in Jhanji River sediments. Sci. Total Envir., 193(1), 1-12.
Borghei SM, Hosseini SH (2004). The treatment of phenolic wastewater using a moving bed biofilm reactor. Proc. Biochem., 39; 1177-1181.
Dubey SK, Hussain A (2014). Phenol biodegradation: a review. Int J Environ Eng., 1(2):151–157.
El-Naggar NE, Abdelwahed NA, Saber WI, Mohamed AA (2014). Bioprocessing of some agro-industrial residues for endoglucanase production by the new subsp.; Streptomyces albogriseolus sub sp. cellulolyticus strain NEAE-J. Brazilian Journal of Microbiology, 45, 2, 743-756.
Fuentes M, Benimeli C, Cuozzo S, Amoroso M (2010). Isolation of pesticide-degrading actinomycetes from a contaminated site: Bacterial growth, removal and dechlorination of organochlorine pesticides. Int. Biodeterior. Biodegrad., 64:434–441.
Gerginova M, Manasiev J, Shivarova N, Alexieva Z (2007). Influence of various phenolic compounds on phenol hydroxylase activity of a Trichosporon cutaneum strain. Z Naturforsch C.; 62(1-2):83-6.
Haritash A and Kaushik C (2009). Biodegradation aspects of polycyclic aromatic hydrocarbons (PAHs): a review. J Hazard Mater., 169:1–15.
Hasan S A and Jabeen S (2015). Degradation kinetics and pathway of phenol by Pseudomonas and Bacillus species. Biotechnology Biotechnological Equipment, 29:1, 45-53
Hussain A, Kumar P, Mehrotra I (2008). Treatment of phenolic wastewater in UASB reactor: efect of Nitrogen and Phosphorous. Biores Technol 99:8497–8503
Hussain A, Kumar P, Mehrotra I (2010). Anaerobic treatment of phenolic wastewater: effect of phosphorous limitation. Desalination Water Treat 20:189–196
Hussain A, Parveen T, Kumar P, Mehrotra I (2009). Phenolic wastewater: effect of F/M on anaerobic degradation. Desalination Water Treat 2:254–259
IARC (1989). International Agency for Research on Cancer “Phenol,” In: IARC monographs on the evaluation of the carcinogenic risk of chemicals to humans, World Health Organization, 47:263-287.
Istianto Y, Koesoemowidodo R, Watanabe Y (2012). Application of Phenol Pretreatment for the Isolation of Rare Actinomycetes from Indonesian Soil . Microbiology Indonesia 6(1):42- 47.
Jaweria R, Peshwe SA, Ingale AG (2013). Biodegradation of P-Nitro Phenol by an Actinomycete. Indian Journal Of Applied Research, 3 (6): 241.
John GH, Sneath PH, Krieg NR, Holt JG, Holt JG (2000). Bergey’s manual of determinative bacteriology, 9th ed. New York, NY: Lippincott Williams and Wilkins.
Khalel A, Alshehri W, Aly M (2020). Enhancing plant growth by chicken feather compost obtained from feather degradation by Streptomyces enissocaesilis. Biosc. Biotec. Res.; 13(4)1847-1853
Khleifat L and Khaled M (2007). Biodegradation of phenol by Actinobacillus sp. Mathematical Interpretation and Effect of some growth conditions. Bioremediation J. 11: 103-112.
Kim BS (2000). Production of poly3-hydroxybutyrate from inexpensive substrates. Enzyme Microbial. Technol., 27: 774-777.
Madan S, Sachan P, Singh U (2018). A review on Bioremediation of Pulp and Paper mill efuent: an alternative to conventional remedial technologies. J Appl Nat Sci., 10(1):367–374.
Malaviya P, Rathore VS (2007). Bioremediation of pulp and paper mill effluent by a novel fungal consortium isolated from polluted soil. Biores Technol., 98:3647–3651
Mallick J, Singh CK, AlMesfer MK, Singh VP and Alsubih M (2021). Groundwater Quality Studies in the Kingdom of Saudi Arabia: Prevalent Research and Management Dimensions. Water, 13, 1266.
Marihal AK, Jagadeesh KS (2013). Plant-microbe interaction: a potential tool for enhanced bioremediation. In: Arora NK (ed) Plant microbe symbiosis: fundamentals and advances. Springer, India, pp 395–410.
Mohd T and Piakong L (2006). The performance of phenol biodegradation by Candida tropicalis RETL-Crl using batch and fed batch Fermentation techniques. Ph.D thesis. University Teknologi Malaysia
Mohite BV, Jalgaonwala RE, Pawar S, Morankar A (2010). Isolation and characterization of phenol degrading bacteria from oil contaminated soil.
Mörsen A and Rehm, HJ (1990). Degradation of phenol by a defined mixed culture immobilized by absorption on activated carbon and sintered glass. Appl. Microbiol. Biotechnol., 33, 209-212.
Myronovskyi M., Rosenkränzer B., Stierhof M., Petzke L., Seiser T., Luzhetskyy A (2020). Identification and heterologous expression of the albucidin gene cluster from the marine strain Streptomyces albus subsp. chlorinus NRRL B- 24108. Microorganisms, 8:237.
Nagamani A., Soligala R., Lowry M (2009). Isolation and characterization of phenol degrading Xanthobacter flavus. African Journal of Biotechnology., 8 (20), 5449-5453
Nair CI, Jayachandran K, Shashidhar S (2008). Biodegradation of phenol. African Journal of Biotechnology, 7 (25), 4951-4958.
Nair IC, Jayachandran K, Shankar S (2007). Treatment of paper factory effluent using a phenol degrading Alcaligenes sp. Under free and immobilized condition. Bioresour. Technol 98: 714-716.
Nakagawa H, Inoue H and Takeda Y (1963). Characteristics of catechol oxygenase from Brevibacterium fuscum. J. Biochem., 54, 65.
Neufeld RD and Poladino SB (1985). Comparision of 4-aminoantipyrine and gas chromatography techniques for analysis of phenol compound. J. Water Pollut. Control Fed. 27(10): 1040-1044
Nilotpala P and Ingle AO (2007). Mineralization of phenol by Serratia plymuthica strain GC isolated from sludge sample. Int. Biodeterior. Biodegradation, 60.2: 103-108.
Park C, Lee M, Lee B, Kim SW, Chase HA, Lee J, Kim S (2007). Biodegradation and biosorption for decolorization of synthetic dyes by Funalia trogii. Biochem Eng J 36:59-65
Pazarlioglu NK and Telefoncu A (2005). Bio degradation of phenol by Pseudomonas putida immobilized on activated pumice particles. Proc Biochem 40:1807–1814
Sachan P, Madan S, Hussain A (2019). Isolation and screening of phenol‑degrading bacteria from pulp and paper mill efuent, Applied Water Science, doi.org/10.1007/s13201-019-0994-9.
Sachan P, Madan S, Hussain A (2019). Isolation and screening of phenol‑degrading bacteria from pulp and paper mill efuent, Applied Water Science, doi.org/10.1007/s13201-019-0994-9
Shawabkeh R, Khaled M, Khleifat I (2007). Rate of biodegradation of phenol by Klebsiella oxytoca in minimal medium and nutrient broth conditions. Bioremediation J., 11: 13-19.
Shazryenna D, Ruzanna R, Jessica MS, Piakong MT (2015). Phenol biodegradation by free and and Immobilized Candida tropicalis RETL-Crl on Coconut Husk and Loofah Packed in Biofilter Column. Conference Series Materials Science and Engineering 78(1) 221-219.
Shirling E. B., Gottlieb D. (1966). Methods for characterization of Streptomyces species. Int. J. Syst. Bacteriol., 16, 313–340.
Singh V, Haque S, Singh H, Verma J, Vibha K, Singh R, Jawed A and Tripathi CK (2016). Isolation, Screening, and Identification of Novel Isolates of Actinomycetes from India for Antimicrobial Applications. Front. Microbiol., 7:1921.
Sun JQ, Xu L, Tang YQ, Chen FM, Wu XL (2012). Simultaneous degradation of phenol and n-hexadecane by Acinetobacter strains. Biores Tech 123:664–668.
Tallur P, Megadi V, Kamanavalli C, Ninnekar H. (2006). Biodegradation of p-Cresol by Bacillus sp. Strain PHN 1. Curr Microbiol.; 53:529-533.
Tork S E, Aly MM, El-semin O (2018). A new l-glutaminase from Streptomyces pratensis NRC 10: Gene identification, enzyme purification, and characterization. International Journal of Biological Macromolecules, 113:550-557.
Tran-Ly AN, Reyes C, Schwarze FR et al. (2020). Microbial production of melanin and its various applications. World J Microbiol Biotechnol., 36: 170.
Weisberg WG, Barns SM, Pelletier DA and Lane DJ (1991). 16S ribosomal DNA amplification for phylogenetic study.J. Bacteriol., 173: 697–703
Xu N, Qiu C, Yang Q, Zhang Y, Wang M, Ye C, Guo M (2021). Analysis of Phenol Biodegradation in Antibiotic and Heavy Metal Resistant Acinetobacter lwoffii NL1. Frontiers in Microbiology, vol 12, P: 2670.
Yang RD and Humphery A.E (1975). Dynamic and steady sate studies of phenol biodegradation in pure and mixed cultures. Biotechnol . Bioeng., 17, 1211-1235.


